7SGW
 
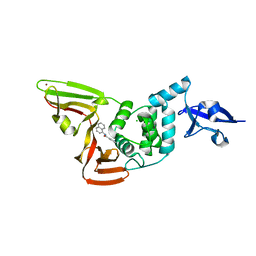 | | Papain-Like Protease of SARS CoV-2 in complex with PLP_Snyder630 inhibitor | | Descriptor: | CHLORIDE ION, N-(naphthalen-1-yl)pyridine-3-carboxamide, Non-structural protein 3, ... | | Authors: | Osipiuk, J, Tesar, C, Endres, M, Lisnyak, V, Maki, S, Taylor, C, Zhang, Y, Zhou, Z, Azizi, S.A, Jones, K, Kathayat, R, Snyder, S.A, Dickinson, B.C, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-10-07 | | Release date: | 2021-10-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Papain-Like Protease of SARS CoV-2 in complex with PLP_Snyder630 inhibitor
To Be Published
|
|
7SGV
 
 | | Papain-Like Protease of SARS CoV-2, C111S mutant, in complex with PLP_Snyder630 inhibitor | | Descriptor: | CHLORIDE ION, N-(naphthalen-1-yl)pyridine-3-carboxamide, Papain-like protease, ... | | Authors: | Osipiuk, J, Tesar, C, Endres, M, Lisnyak, V, Maki, S, Taylor, C, Zhang, Y, Zhou, Z, Azizi, S.A, Jones, K, Kathayat, R, Snyder, S.A, Dickinson, B.C, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-10-07 | | Release date: | 2021-10-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Papain-Like Protease of SARS CoV-2, C111S mutant, in complex with PLP_Snyder630 inhibitor
To Be Published
|
|
7SGU
 
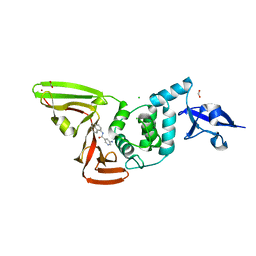 | | Papain-Like Protease of SARS CoV-2, C111S mutant, in complex with PLP_Snyder608 inhibitor | | Descriptor: | 5-amino-N-(naphthalen-1-yl)pyridine-3-carboxamide, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Osipiuk, J, Tesar, C, Endres, M, Lisnyak, V, Maki, S, Taylor, C, Zhang, Y, Zhou, Z, Azizi, S.A, Jones, K, Kathayat, R, Snyder, S.A, Dickinson, B.C, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-10-07 | | Release date: | 2021-10-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Papain-Like Protease of SARS CoV-2, C111S mutant, in complex with PLP_Snyder608 inhibitor
To Be Published
|
|
7JN2
 
 | | The crystal structure of Papain-Like Protease of SARS CoV-2 in complex with PLP_Snyder441 inhibitor | | Descriptor: | 3-amino-2-methyl-N-[(1R)-1-(naphthalen-1-yl)ethyl]benzamide, ACETATE ION, CHLORIDE ION, ... | | Authors: | Osipiuk, J, Tesar, C, Endres, M, Lisnyak, V, Maki, S, Taylor, C, Zhang, Y, Zhou, Z, Azizi, S.A, Jones, K, Kathayat, R, Snyder, S.A, Dickinson, B.C, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-08-03 | | Release date: | 2020-08-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | The crystal structure of Papain-Like Protease of SARS CoV-2 in complex with PLP_Snyder441
to be published
|
|
7KOJ
 
 | | The crystal structure of Papain-Like Protease of SARS CoV-2, C111S mutant, in complex with PLP_Snyder494 inhibitor | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-methyl-N-[(1R)-1-(naphthalen-1-yl)ethyl]-5-{[(prop-2-en-1-yl)carbamoyl]amino}benzamide, ACETATE ION, ... | | Authors: | Osipiuk, J, Tesar, C, Endres, M, Lisnyak, V, Maki, S, Taylor, C, Zhang, Y, Zhou, Z, Azizi, S.A, Jones, K, Kathayat, R, Snyder, S.A, Dickinson, B.C, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-11-09 | | Release date: | 2020-11-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | The crystal structure of Papain-Like Protease of SARS CoV-2, C111S mutant, in complex with PLP_Snyder494
to be published
|
|
7KOL
 
 | | The crystal structure of Papain-Like Protease of SARS CoV-2 in complex with PLP_Snyder496 inhibitor | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5-[(E)-(hydroxyimino)methyl]-2-methyl-N-[(1R)-1-(naphthalen-1-yl)ethyl]benzamide, CHLORIDE ION, ... | | Authors: | Osipiuk, J, Tesar, C, Endres, M, Lisnyak, V, Maki, S, Taylor, C, Zhang, Y, Zhou, Z, Azizi, S.A, Jones, K, Kathayat, R, Snyder, S.A, Dickinson, B.C, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-11-09 | | Release date: | 2020-11-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | The crystal structure of Papain-Like Protease of SARS CoV-2 in complex with PLP_Snyder496
to be published
|
|
7KOK
 
 | | The crystal structure of Papain-Like Protease of SARS CoV-2, C111S mutant, in complex with PLP_Snyder496 inhibitor | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5-[(E)-(hydroxyimino)methyl]-2-methyl-N-[(1R)-1-(naphthalen-1-yl)ethyl]benzamide, ACETATE ION, ... | | Authors: | Osipiuk, J, Tesar, C, Endres, M, Lisnyak, V, Maki, S, Taylor, C, Zhang, Y, Zhou, Z, Azizi, S.A, Jones, K, Kathayat, R, Snyder, S.A, Dickinson, B.C, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-11-09 | | Release date: | 2020-11-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of Papain-Like Protease of SARS CoV-2, C111S mutant, in complex with PLP_Snyder496
to be published
|
|
7KRX
 
 | | The crystal structure of Papain-Like Protease of SARS CoV-2, C111S mutant, in complex with PLP_Snyder441 inhibitor | | Descriptor: | 3-amino-2-methyl-N-[(1R)-1-(naphthalen-1-yl)ethyl]benzamide, ACETATE ION, CHLORIDE ION, ... | | Authors: | Osipiuk, J, Tesar, C, Endres, M, Lisnyak, V, Maki, S, Taylor, C, Zhang, Y, Zhou, Z, Azizi, S.A, Jones, K, Kathayat, R, Snyder, S.A, Dickinson, B.C, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-11-20 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | The crystal structure of Papain-Like Protease of SARS CoV-2, C111S mutant, in complex with PLP_Snyder441
to be published
|
|
7QB2
 
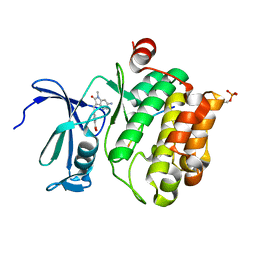 | | Pim1 in complex with (E)-4-((6-amino-1-methyl-2-oxoindolin-3-ylidene)methyl)benzoic acid and Pimtide | | Descriptor: | 4-[(E)-(6-azanyl-1-methyl-2-oxidanylidene-indol-3-ylidene)methyl]benzoic acid, GLYCEROL, Pimtide, ... | | Authors: | Hochban, P.M.M, Heine, A, Diederich, W.E. | | Deposit date: | 2021-11-18 | | Release date: | 2022-11-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Pose, duplicate, then elaborate: Steps towards increased affinity for inhibitors targeting the specificity surface of the Pim-1 kinase.
Eur.J.Med.Chem., 245, 2023
|
|
7QFM
 
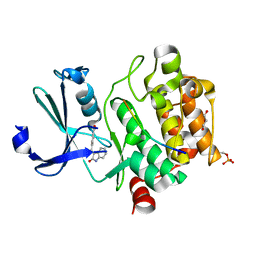 | | Pim1 in complex with (E)-4-((2-oxoindolin-3-ylidene)methyl)benzoic acid and Pimtide | | Descriptor: | 4-[(~{E})-(2-oxidanylidene-1~{H}-indol-3-ylidene)methyl]benzoic acid, GLYCEROL, Pimtide, ... | | Authors: | Hochban, P.M.M, Heine, A, Diederich, W.E. | | Deposit date: | 2021-12-06 | | Release date: | 2022-12-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Pose, duplicate, then elaborate: Steps towards increased affinity for inhibitors targeting the specificity surface of the Pim-1 kinase.
Eur.J.Med.Chem., 245, 2023
|
|
7Z6U
 
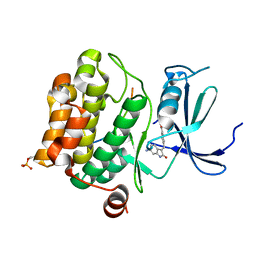 | | Pim1 in complex with (E)-4-((6-amino-2-oxoindolin-3-ylidene)methyl)benzoic acid and Pimtide | | Descriptor: | 4-[(~{E})-(6-azanyl-2-oxidanylidene-1~{H}-indol-3-ylidene)methyl]benzoic acid, GLYCEROL, Isoform 1 of Serine/threonine-protein kinase pim-1, ... | | Authors: | Hochban, P.M.M, Heine, A, Diederich, W.E. | | Deposit date: | 2022-03-14 | | Release date: | 2023-02-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Pose, duplicate, then elaborate: Steps towards increased affinity for inhibitors targeting the specificity surface of the Pim-1 kinase.
Eur.J.Med.Chem., 245, 2023
|
|
8AFR
 
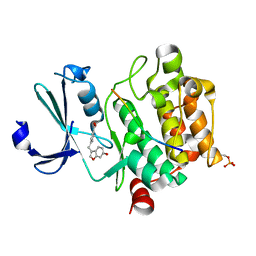 | | Pim1 in complex with 4-((6-hydroxybenzofuran-3-yl)methyl)benzoic acid and Pimtide | | Descriptor: | 4-((6-hydroxybenzofuran-3-yl)methyl)benzoic acid, Pimtide, Serine/threonine-protein kinase pim-1 | | Authors: | Hochban, P.M.M, Heine, A, Diederich, W.E. | | Deposit date: | 2022-07-18 | | Release date: | 2023-02-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Pose, duplicate, then elaborate: Steps towards increased affinity for inhibitors targeting the specificity surface of the Pim-1 kinase.
Eur.J.Med.Chem., 245, 2023
|
|
5MLE
 
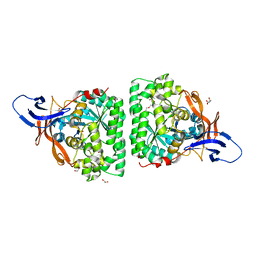 | | Crystal Structure of Human Dihydropyrimidinease-like 2 (DPYSL2A)/Collapsin Response Mediator Protein (CRMP2 13-516) Mutant Y479E/Y499E | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Dihydropyrimidinase-related protein 2, ... | | Authors: | Sethi, R, Zheng, Y, Talon, R, Velupillai, S, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Ahmed, A.A, von Delft, F. | | Deposit date: | 2016-12-06 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Tuning microtubule dynamics to enhance cancer therapy by modulating FER-mediated CRMP2 phosphorylation.
Nat Commun, 9, 2018
|
|
5MKV
 
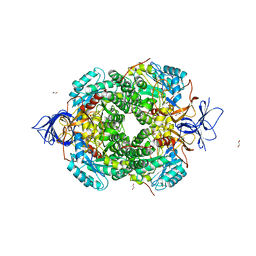 | | Crystal Structure of Human Dihydropyrimidinease-like 2 (DPYSL2A)/Collapsin Response Mediator Protein (CRMP2) residues 13-516 | | Descriptor: | 1,2-ETHANEDIOL, Dihydropyrimidinase-related protein 2 | | Authors: | Sethi, R, Zheng, Y, Krojer, T, Velupillai, S, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Ahmed, A.A, von Delft, F. | | Deposit date: | 2016-12-05 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Tuning microtubule dynamics to enhance cancer therapy by modulating FER-mediated CRMP2 phosphorylation.
Nat Commun, 9, 2018
|
|
4F3L
 
 | | Crystal Structure of the Heterodimeric CLOCK:BMAL1 Transcriptional Activator Complex | | Descriptor: | BMAL1b, Circadian locomoter output cycles protein kaput | | Authors: | Huang, N, Chelliah, Y, Shan, Y, Taylor, C, Yoo, S, Partch, C, Green, C.B, Zhang, H, Takahashi, J. | | Deposit date: | 2012-05-09 | | Release date: | 2012-06-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.268 Å) | | Cite: | Crystal structure of the heterodimeric CLOCK:BMAL1 transcriptional activator complex.
Science, 337, 2012
|
|
7MJR
 
 | | Vip4Da2 toxin structure | | Descriptor: | CALCIUM ION, SULFATE ION, Vip4Da1 protein | | Authors: | Rydel, T.J, Duda, D, Zheng, M, Henry, A. | | Deposit date: | 2021-04-20 | | Release date: | 2021-05-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.22 Å) | | Cite: | Structural and functional insights into the first Bacillus thuringiensis vegetative insecticidal protein of the Vpb4 fold, active against western corn rootworm.
Plos One, 16, 2021
|
|
5N5M
 
 | |
5N5L
 
 | |
6YND
 
 | | GAPDH purified from the supernatant of HEK293F cells: crystal form 1 of 4. | | Descriptor: | 3,6,9,12,15,18,21,24,27-NONAOXANONACOSANE-1,29-DIOL, Glyceraldehyde-3-phosphate dehydrogenase | | Authors: | Roversi, P, Lia, A. | | Deposit date: | 2020-04-13 | | Release date: | 2020-05-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.525 Å) | | Cite: | Partial catalytic Cys oxidation of human GAPDH to Cys-sulfonic acid.
Wellcome Open Res, 5, 2020
|
|
6YNF
 
 | | GAPDH purified from the supernatant of HEK293F cells: crystal form 3 of 4. | | Descriptor: | 3,6,9,12,15,18,21,24,27-NONAOXANONACOSANE-1,29-DIOL, Glyceraldehyde-3-phosphate dehydrogenase | | Authors: | Roversi, P, Lia, A. | | Deposit date: | 2020-04-13 | | Release date: | 2020-05-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.394 Å) | | Cite: | Partial catalytic Cys oxidation of human GAPDH to Cys-sulfonic acid.
Wellcome Open Res, 5, 2020
|
|
6YNE
 
 | | GAPDH purified from the supernatant of HEK293F cells: crystal form 2 of 4. | | Descriptor: | 3,6,9,12,15,18,21,24,27-NONAOXANONACOSANE-1,29-DIOL, Glyceraldehyde-3-phosphate dehydrogenase | | Authors: | Roversi, P, Lia, A. | | Deposit date: | 2020-04-13 | | Release date: | 2020-05-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.853 Å) | | Cite: | Partial catalytic Cys oxidation of human GAPDH to Cys-sulfonic acid.
Wellcome Open Res, 5, 2020
|
|
6YNH
 
 | | GAPDH purified from the supernatant of HEK293F cells: crystal form 4 of 4 | | Descriptor: | 3,6,9,12,15,18,21,24,27-NONAOXANONACOSANE-1,29-DIOL, Glyceraldehyde-3-phosphate dehydrogenase | | Authors: | Roversi, P, Lia, A. | | Deposit date: | 2020-04-13 | | Release date: | 2020-05-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.621 Å) | | Cite: | Partial catalytic Cys oxidation of human GAPDH to Cys-sulfonic acid.
Wellcome Open Res, 5, 2020
|
|
5NAF
 
 | | Co-crystal structure of an MeCP2 peptide with TBLR1 WD40 domain | | Descriptor: | F-box-like/WD repeat-containing protein TBL1XR1, GLYCEROL, Methyl-CpG-binding protein 2 | | Authors: | Kruusvee, V, Cook, A.G. | | Deposit date: | 2017-02-27 | | Release date: | 2017-03-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.493 Å) | | Cite: | Structure of the MeCP2-TBLR1 complex reveals a molecular basis for Rett syndrome and related disorders.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7JIT
 
 | | The crystal structure of Papain-Like Protease of SARS CoV-2 , C111S mutant, in complex with PLP_Snyder495 inhibitor | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5-[(carbamoylcarbamoyl)amino]-2-methyl-N-[(1R)-1-(naphthalen-1-yl)ethyl]benzamide, ACETATE ION, ... | | Authors: | Osipiuk, J, Tesar, C, Endres, M, Lisnyak, V, Maki, S, Taylor, C, Zhang, Y, Zhou, Z, Azizi, S.A, Jones, K, Kathayat, R, Snyder, S.A, Dickinson, B.C, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-07-23 | | Release date: | 2020-08-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of papain-like protease from SARS-CoV-2 and its complexes with non-covalent inhibitors.
Nat Commun, 12, 2021
|
|
7JIR
 
 | | The crystal structure of Papain-Like Protease of SARS CoV-2 , C111S mutant, in complex with PLP_Snyder457 inhibitor | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5-amino-2-methyl-N-[(1R)-1-naphthalen-1-ylethyl]benzamide, ACETATE ION, ... | | Authors: | Osipiuk, J, Tesar, C, Endres, M, Lisnyak, V, Maki, S, Taylor, C, Zhang, Y, Zhou, Z, Azizi, S.A, Jones, K, Kathayat, R, Snyder, S.A, Dickinson, B.C, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-07-23 | | Release date: | 2020-08-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structure of papain-like protease from SARS-CoV-2 and its complexes with non-covalent inhibitors.
Nat Commun, 12, 2021
|
|
