3JYC
 
 | |
6V3G
 
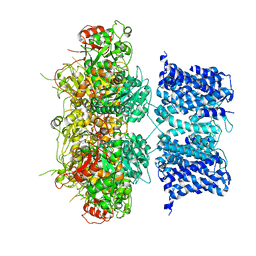 | | Cryo-EM structure of Ca2+-free hsSlo1 channel | | Descriptor: | Calcium-activated potassium channel subunit alpha-1 | | Authors: | Tao, X, MacKinnon, R. | | Deposit date: | 2019-11-25 | | Release date: | 2019-12-25 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Molecular structures of the human Slo1 K + channel in complex with beta 4.
Elife, 8, 2019
|
|
1LXN
 
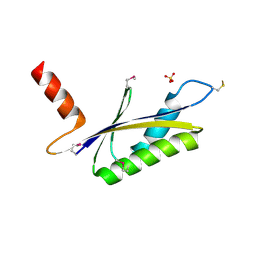 | | X-RAY STRUCTURE OF MTH1187 NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET TT272 | | Descriptor: | HYPOTHETICAL PROTEIN MTH1187, SULFATE ION | | Authors: | Tao, X, Khayat, R, Christendat, D, Savchenko, A, Xu, X, Edwards, A, Arrowsmith, C.H, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2002-06-05 | | Release date: | 2003-07-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of MTH1187 and its Yeast Ortholog YBL001C
Proteins, 52, 2003
|
|
1LXJ
 
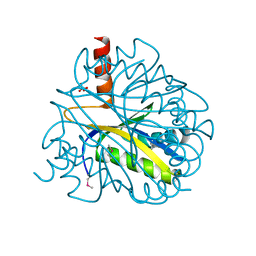 | | X-RAY STRUCTURE OF YBL001c NORTHEAST STRUCTURAL GENOMICS (NESG) CONSORTIUM TARGET YTYst72 | | Descriptor: | HYPOTHETICAL 11.5KDA PROTEIN IN HTB2-NTH2 INTERGENIC REGION, SULFATE ION | | Authors: | Tao, X, Khayat, R, Christendat, D, Savchenko, A, Xu, X, Edwards, A, Arrowsmith, C.H, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2002-06-05 | | Release date: | 2003-07-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | CRYSTAL STRUCTURES OF MTH1187 AND ITS YEAST ORTHOLOG YBL001C
Proteins, 52, 2003
|
|
3LNM
 
 | | F233W mutant of the Kv2.1 paddle-Kv1.2 chimera channel | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, F233W mutant of the Kv2.1 paddle-Kv1.2 chimera, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Tao, X, Lee, A, Limapichat, W, Dougherty, D.A, MacKinnon, R. | | Deposit date: | 2010-02-02 | | Release date: | 2010-04-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A gating charge transfer center in voltage sensors.
Science, 328, 2010
|
|
1PJ2
 
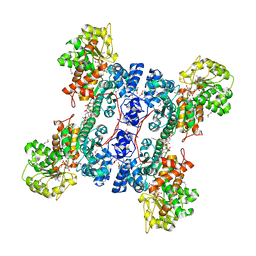 | | Crystal structure of human mitochondrial NAD(P)+-dependent malic enzyme in a pentary complex with natural substrate malate, cofactor NADH, Mn++, and allosteric activator fumarate | | Descriptor: | (2S)-2-hydroxybutanedioic acid, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, FUMARIC ACID, ... | | Authors: | Tao, X, Yang, Z, Tong, L. | | Deposit date: | 2003-05-30 | | Release date: | 2003-11-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of substrate complexes of malic enzyme and insights into the catalytic mechanism.
Structure, 11, 2003
|
|
1PJ4
 
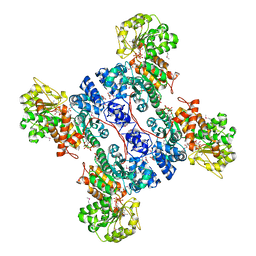 | | Crystal structure of human mitochondrial NAD(P)+-dependent malic enzyme in a pentary complex with natural substrate malate, ATP, Mn++, and allosteric activator fumarate. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, D-MALATE, FUMARIC ACID, ... | | Authors: | Tao, X, Yang, Z, Tong, L. | | Deposit date: | 2003-05-31 | | Release date: | 2003-09-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of substrate complexes of malic enzyme and insights into the catalytic mechanism.
Structure, 11, 2003
|
|
1O77
 
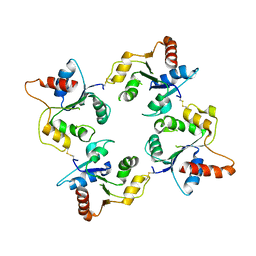 | | CRYSTAL STRUCTURE OF THE C713S MUTANT OF THE TIR DOMAIN OF HUMAN TLR2 | | Descriptor: | TOLL-LIKE RECEPTOR 2 | | Authors: | Tao, X, Xu, Y, Ye, Z, Beg, A.A, Tong, L. | | Deposit date: | 2002-10-24 | | Release date: | 2002-11-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | An Extensively Associated Dimer in the Structure of the C713S Mutant of the Tir Domain of Human Tlr2
Biochem.Biophys.Res.Commun., 299, 2002
|
|
1PJ3
 
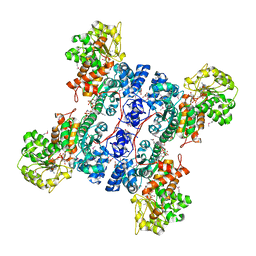 | | Crystal structure of human mitochondrial NAD(P)+-dependent malic enzyme in a pentary complex with natural substrate pyruvate, cofactor NAD+, Mn++, and allosteric activator fumarate. | | Descriptor: | FUMARIC ACID, MANGANESE (II) ION, NAD-dependent malic enzyme, ... | | Authors: | Tao, X, Yang, Z, Tong, L. | | Deposit date: | 2003-05-30 | | Release date: | 2003-11-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of substrate complexes of malic enzyme and insights into the catalytic mechanism.
Structure, 11, 2003
|
|
1PE0
 
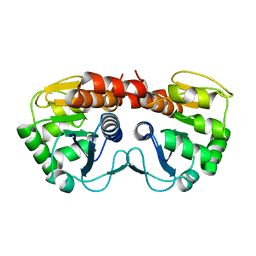 | |
1PDV
 
 | |
1PDW
 
 | |
2OUD
 
 | | Crystal structure of the catalytic domain of human MKP5 | | Descriptor: | CHLORIDE ION, Dual specificity protein phosphatase 10 | | Authors: | Tao, X, Tong, L. | | Deposit date: | 2007-02-10 | | Release date: | 2007-08-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the MAP kinase binding domain and the catalytic domain of human MKP5.
Protein Sci., 16, 2007
|
|
2OUC
 
 | |
8GHF
 
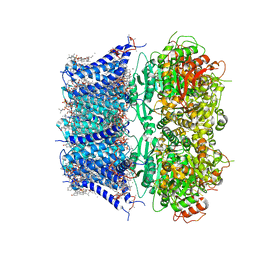 | | cryo-EM structure of hSlo1 in plasma membrane vesicles | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, CALCIUM ION, CHOLESTEROL, ... | | Authors: | Tao, X, Zhao, C, MacKinnon, R. | | Deposit date: | 2023-03-10 | | Release date: | 2023-05-10 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Membrane protein isolation and structure determination in cell-derived membrane vesicles.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8GHG
 
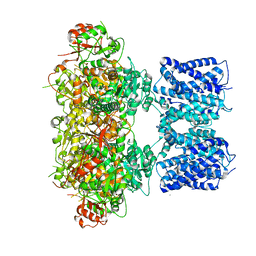 | |
8GH9
 
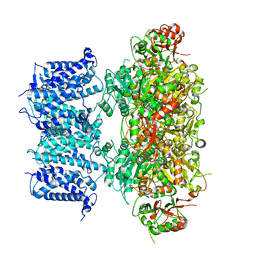 | |
8XJW
 
 | | Structure of the Argonaute protein from Kurthia massiliensis in complex with guide DNA and 19-mer target RNA | | Descriptor: | KmAgo, MANGANESE (II) ION, guide DNA, ... | | Authors: | Tao, X, Ding, H, Wu, S. | | Deposit date: | 2023-12-22 | | Release date: | 2024-12-18 | | Last modified: | 2025-07-16 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Structural and mechanistic insights into a mesophilic prokaryotic Argonaute.
Nucleic Acids Res., 52, 2024
|
|
8XHS
 
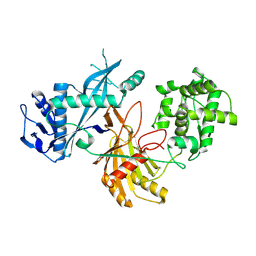 | | Cryo-EM structure of free-state KmAgo | | Descriptor: | KmAgo | | Authors: | Tao, X, Ding, H, Wu, S. | | Deposit date: | 2023-12-18 | | Release date: | 2024-12-25 | | Last modified: | 2025-07-16 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Structural and mechanistic insights into a mesophilic prokaryotic Argonaute.
Nucleic Acids Res., 52, 2024
|
|
8XK4
 
 | | Structure of the Argonaute protein from Kurthia massiliensis in complex with guide DNA and 19-mer target DNA in target-released state | | Descriptor: | 2'-DEOXYCYTIDINE-5'-MONOPHOSPHATE, KmAgo, MANGANESE (II) ION, ... | | Authors: | Tao, X, Ding, H, Wu, S. | | Deposit date: | 2023-12-22 | | Release date: | 2025-01-01 | | Last modified: | 2025-07-16 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Structural and mechanistic insights into a mesophilic prokaryotic Argonaute.
Nucleic Acids Res., 52, 2024
|
|
8XHV
 
 | |
8XK0
 
 | | Structure of the Argonaute protein from Kurthia massiliensis in complex with guide DNA and 19-mer target DNA in pre-cleavage state | | Descriptor: | KmAgo, MANGANESE (II) ION, guide DNA, ... | | Authors: | Tao, X, Ding, H, Wu, S. | | Deposit date: | 2023-12-22 | | Release date: | 2024-12-18 | | Last modified: | 2025-07-16 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Structural and mechanistic insights into a mesophilic prokaryotic Argonaute.
Nucleic Acids Res., 52, 2024
|
|
8XJX
 
 | | Structure of the Argonaute protein from Kurthia massiliensis in complex with guide DNA and 19-mer target DNA | | Descriptor: | KmAgo, MANGANESE (II) ION, guide DNA, ... | | Authors: | Tao, X, Ding, H, Wu, S. | | Deposit date: | 2023-12-22 | | Release date: | 2025-01-01 | | Last modified: | 2025-07-16 | | Method: | ELECTRON MICROSCOPY (2.78 Å) | | Cite: | Structural and mechanistic insights into a mesophilic prokaryotic Argonaute.
Nucleic Acids Res., 52, 2024
|
|
8XK3
 
 | | Structure of the Argonaute protein from Kurthia massiliensis in complex with guide DNA and 19-mer target DNA in target-cleaved state | | Descriptor: | KmAgo, MANGANESE (II) ION, guide DNA, ... | | Authors: | Tao, X, Ding, H, Wu, S. | | Deposit date: | 2023-12-22 | | Release date: | 2025-01-01 | | Last modified: | 2025-07-16 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Structural and mechanistic insights into a mesophilic prokaryotic Argonaute.
Nucleic Acids Res., 52, 2024
|
|
6UWM
 
 | | Single particle cryo-EM structure of KvAP | | Descriptor: | Voltage-gated potassium channel | | Authors: | Tao, X, MacKinnon, R. | | Deposit date: | 2019-11-05 | | Release date: | 2019-12-04 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | Cryo-EM structure of the KvAP channel reveals a non-domain-swapped voltage sensor topology.
Elife, 8, 2019
|
|
