3FSS
 
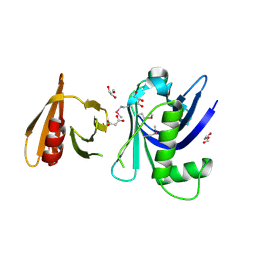 | | Structure of the tandem PH domains of Rtt106 | | Descriptor: | GLYCEROL, Histone chaperone RTT106, MALONIC ACID | | Authors: | Su, D, Thompson, J.R, Mer, G. | | Deposit date: | 2009-01-11 | | Release date: | 2009-12-22 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.432 Å) | | Cite: | Structural basis for recognition of H3K56-acetylated histone H3-H4 by the chaperone Rtt106.
Nature, 483, 2012
|
|
2GA6
 
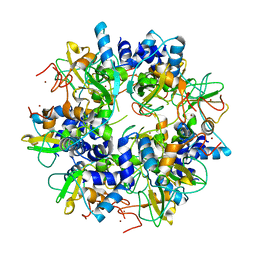 | | The crystal structure of SARS nsp10 without zinc ion as additive | | Descriptor: | ZINC ION, orf1a polyprotein | | Authors: | Su, D, Lou, Z, Sun, F, Zhai, Y, Yang, H, Rao, Z. | | Deposit date: | 2006-03-08 | | Release date: | 2006-08-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Dodecamer Structure of Severe Acute Respiratory Syndrome Coronavirus Nonstructural Protein nsp10
J.Virol., 80, 2006
|
|
2G9T
 
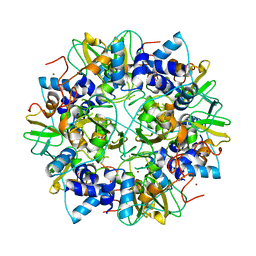 | | Crystal structure of the SARS coronavirus nsp10 at 2.1A | | Descriptor: | ZINC ION, orf1a polyprotein | | Authors: | Su, D, Lou, Z, Yang, H, Sun, F, Rao, Z. | | Deposit date: | 2006-03-07 | | Release date: | 2006-08-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Dodecamer Structure of Severe Acute Respiratory Syndrome Coronavirus Nonstructural Protein nsp10
J.Virol., 80, 2006
|
|
8IH7
 
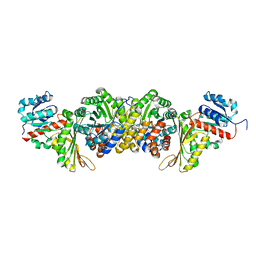 | | AmnG-AmnH complex | | Descriptor: | 4-hydroxy-2-oxovalerate aldolase, Acetaldehyde dehydrogenase, OXALATE ION, ... | | Authors: | Su, D, Shi, Q.L. | | Deposit date: | 2023-02-22 | | Release date: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | AmnG-AmnH complex
To Be Published
|
|
3TW1
 
 | | Structure of Rtt106-AHN | | Descriptor: | GLYCEROL, Histone chaperone RTT106, N-[2-(1H-IMIDAZOL-4-YL)ETHYL]ACETAMIDE | | Authors: | Su, D, Thompson, J.R, Mer, G. | | Deposit date: | 2011-09-21 | | Release date: | 2012-02-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.772 Å) | | Cite: | Structural basis for recognition of H3K56-acetylated histone H3-H4 by the chaperone Rtt106.
Nature, 483, 2012
|
|
3TVV
 
 | |
3Q66
 
 | |
3Q68
 
 | |
5UMT
 
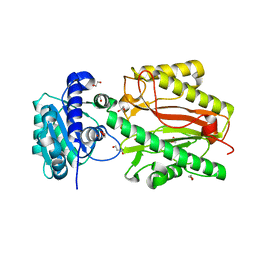 | | Crystal structure of N-terminal domain of human FACT complex subunit SPT16 | | Descriptor: | 1,2-ETHANEDIOL, FACT complex subunit SPT16, GLYCEROL | | Authors: | Su, D, Hu, Q, Thompson, J.R, Botuyan, M.V, Mer, G. | | Deposit date: | 2017-01-29 | | Release date: | 2018-01-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.092 Å) | | Cite: | Crystal structure of N-terminal domain of human FACT complex subunit SPT16
To Be Published
|
|
7BXW
 
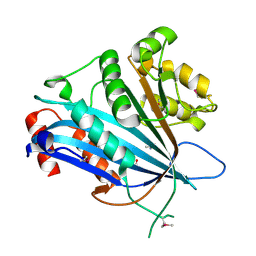 | |
5UMS
 
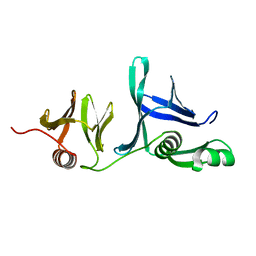 | |
5UMR
 
 | | Crystal structure of N-terminal domain of human FACT complex subunit SSRP1 | | Descriptor: | FACT complex subunit SSRP1 | | Authors: | Su, D, Hu, Q, Thompson, J.R, Heroux, A, Botuyan, M.V, Mer, G. | | Deposit date: | 2017-01-29 | | Release date: | 2018-01-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | Crystal structure of N-terminal domain of human FACT complex subunit SSRP1
To Be Published
|
|
5XM2
 
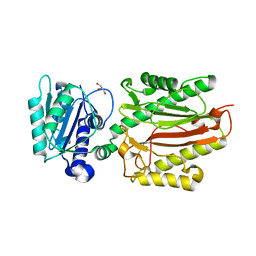 | | Human N-terminal domain of FACT complex subunit SPT16 | | Descriptor: | DI(HYDROXYETHYL)ETHER, FACT complex subunit SPT16, GLYCEROL | | Authors: | Xu, S, Li, H, Dou, Y, Chen, Y, Jiang, H, Lu, D, Wang, M, Su, D. | | Deposit date: | 2017-05-12 | | Release date: | 2018-05-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.187 Å) | | Cite: | The structural basis of human Spt16 N-terminal domain interaction with histone (H3-H4)2tetramer.
Biochem.Biophys.Res.Commun., 508, 2019
|
|
6L4R
 
 | | Crystal structure of Enterovirus D68 RdRp | | Descriptor: | RdRp | | Authors: | Wang, M.L, Li, L, Zhang, Y, Chen, Y.P, Su, D. | | Deposit date: | 2019-10-21 | | Release date: | 2020-06-10 | | Last modified: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (2.147 Å) | | Cite: | Structure of the enterovirus D68 RNA-dependent RNA polymerase in complex with NADPH implicates an inhibitor binding site in the RNA template tunnel.
J.Struct.Biol., 211, 2020
|
|
6L1R
 
 | |
6L1E
 
 | |
6L34
 
 | |
5C94
 
 | | Infectious bronchitis virus nsp9 | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Non-structural protein 9 | | Authors: | Chen, C, Dou, Y, Yang, H, Su, D. | | Deposit date: | 2015-06-26 | | Release date: | 2016-06-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.438 Å) | | Cite: | Structural basis for dimerization and RNA binding of avian infectious bronchitis virus nsp9.
Protein Sci., 26, 2017
|
|
1ZOY
 
 | | Crystal Structure of Mitochondrial Respiratory Complex II from porcine heart at 2.4 Angstroms | | Descriptor: | FAD-binding protein, FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, ... | | Authors: | Sun, F, Huo, X, Zhai, Y, Wang, A, Xu, J, Su, D, Bartlam, M, Rao, Z. | | Deposit date: | 2005-05-15 | | Release date: | 2005-07-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Mitochondrial Respiratory Membrane Protein Complex II
Cell(Cambridge,Mass.), 121, 2005
|
|
1ZP0
 
 | | Crystal Structure of Mitochondrial Respiratory Complex II bound with 3-nitropropionate and 2-thenoyltrifluoroacetone | | Descriptor: | 3-NITROPROPANOIC ACID, 4,4,4-TRIFLUORO-1-THIEN-2-YLBUTANE-1,3-DIONE, FAD-binding protein, ... | | Authors: | Sun, F, Huo, X, Zhai, Y, Wang, A, Xu, J, Su, D, Bartlam, M, Rao, Z. | | Deposit date: | 2005-05-16 | | Release date: | 2005-07-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal Structure of Mitochondrial Respiratory Membrane Protein Complex II
Cell(Cambridge,Mass.), 121, 2005
|
|
5TJB
 
 | | I-II linker of TRPML1 channel at pH 4.5 | | Descriptor: | Mucolipin-1 | | Authors: | Li, M, Zhang, W.K, Benvin, N.M, Zhou, X, Su, D, Li, H, Wang, S, Michailidis, I.E, Tong, L, Li, X, Yang, J. | | Deposit date: | 2016-10-04 | | Release date: | 2017-01-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of dual Ca(2+)/pH regulation of the endolysosomal TRPML1 channel.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5TJA
 
 | | I-II linker of TRPML1 channel at pH 6 | | Descriptor: | Mucolipin-1 | | Authors: | Li, M, Zhang, W.K, Benvin, N.M, Zhou, X, Su, D, Li, H, Wang, S, Michailidis, I.E, Tong, L, Li, X, Yang, J. | | Deposit date: | 2016-10-04 | | Release date: | 2017-01-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of dual Ca(2+)/pH regulation of the endolysosomal TRPML1 channel.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5TJC
 
 | | I-II linker of TRPML1 channel at pH 7.5 | | Descriptor: | Mucolipin-1 | | Authors: | Li, M, Zhang, W.K, Benvin, N.M, Zhou, X, Su, D, Li, H, Wang, S, Michailidis, I.E, Tong, L, Li, X, Yang, J. | | Deposit date: | 2016-10-04 | | Release date: | 2017-01-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of dual Ca(2+)/pH regulation of the endolysosomal TRPML1 channel.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5YPS
 
 | | The structural basis of histone chaperoneVps75 | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Chen, Y, Zhang, Y, Dou, Y, Wang, M, Xu, S, Jiang, H, Limper, A, Su, D. | | Deposit date: | 2017-11-03 | | Release date: | 2018-11-07 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.097 Å) | | Cite: | Structural basis for the acetylation of histone H3K9 and H3K27 mediated by the histone chaperone Vps75 inPneumocystis carinii.
Signal Transduct Target Ther, 4, 2019
|
|
5YYC
 
 | | Crystal structure of alanine racemase from Bacillus pseudofirmus (OF4) | | Descriptor: | Alanine racemase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Dong, H, Hu, T.T, He, G.Z, Lu, D.R, Qi, J.X, Dou, Y.S, Long, W, He, X, Su, D, Ju, J.S. | | Deposit date: | 2017-12-08 | | Release date: | 2019-01-02 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Structural features and kinetic characterization of alanine racemase from Bacillus pseudofirmus OF4.
Biochem. Biophys. Res. Commun., 497, 2018
|
|
