3QUM
 
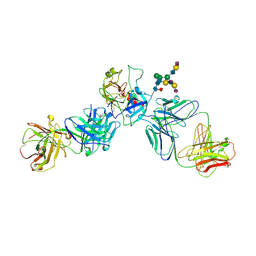 | | Crystal structure of human prostate specific antigen (PSA) in Fab sandwich with a high affinity and a PCa selective antibody | | Descriptor: | Fab 5D3D11 Heavy Chain, Fab 5D3D11 Light Chain, Fab 5D5A5 Heavy Chain, ... | | Authors: | Stura, E.A, Muller, B.H, Michel, S, Ducancel, F. | | Deposit date: | 2011-02-24 | | Release date: | 2011-11-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of human prostate-specific antigen in a sandwich antibody complex.
J.Mol.Biol., 414, 2011
|
|
2C8G
 
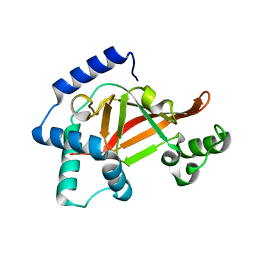 | | Structure of the PN loop Q182A mutant C3bot1 Exoenzyme (Free state, crystal form I) | | Descriptor: | MONO-ADP-RIBOSYLTRANSFERASE C3, SULFATE ION | | Authors: | Stura, E.A, Menetrey, J, Flatau, G, Boquet, P, Menez, A. | | Deposit date: | 2005-12-03 | | Release date: | 2007-02-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Properties of Wild-Type and Two Artt Motif Mutants Clostridium Botulinum C3 Exoenzyme Isoform 1 in Different Substrate Complexed States and Crystal Forms.
To be Published
|
|
2C8H
 
 | | Structure of the PN loop Q182A mutant C3bot1 Exoenzyme (NAD-bound state, crystal form I) | | Descriptor: | MONO-ADP-RIBOSYLTRANSFERASE C3, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION | | Authors: | Stura, E.A, Menetrey, J, Flatau, G, Boquet, P, Menez, A. | | Deposit date: | 2005-12-03 | | Release date: | 2007-02-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural Properties of Wild-Type and Two Artt Motif Mutants Clostridium Botulinum C3 Exoenzyme Isoform 1 in Different Substrate Complexed States and Crystal Forms.
To be Published
|
|
4ELC
 
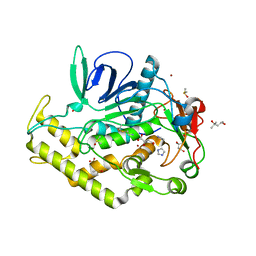 | | Crystal structure of the catalytic domain of botulinum neurotoxin BoNT/A C134 mutant with MTSEA modified Cys-165 | | Descriptor: | (2S)-2-hydroxybutanedioic acid, (4S)-2-METHYL-2,4-PENTANEDIOL, Botulinum neurotoxin A light chain, ... | | Authors: | Stura, E.A, Vera, L, Ptchelkine, D, Bakirci, H, Garcia, S, Dive, V. | | Deposit date: | 2012-04-10 | | Release date: | 2012-08-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Framework for Covalent Inhibition of Clostridium botulinum Neurotoxin A by Targeting Cys165.
J.Biol.Chem., 287, 2012
|
|
4FVL
 
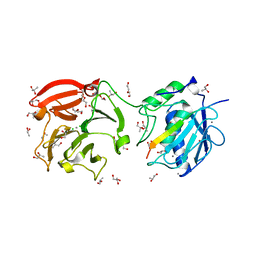 | | Human collagenase 3 (MMP-13) full form with peptides from pro-domain | | Descriptor: | CALCIUM ION, CHLORIDE ION, Collagenase 3, ... | | Authors: | Stura, E.A, Vera, L, Visse, R, Nagase, H, Dive, V. | | Deposit date: | 2012-06-29 | | Release date: | 2013-08-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.436 Å) | | Cite: | Crystal structure of full-length human collagenase 3 (MMP-13) with peptides in the active site defines exosites in the catalytic domain.
Faseb J., 27, 2013
|
|
4G0D
 
 | | Human collagenase 3 (MMP-13) full form with peptides from pro-domain | | Descriptor: | CALCIUM ION, CHLORIDE ION, Collagenase 3, ... | | Authors: | Stura, E.A, Vera, L, Visse, R, Nagase, H, Dive, V. | | Deposit date: | 2012-07-09 | | Release date: | 2013-08-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Crystal structure of full-length human collagenase 3 (MMP-13) with peptides in the active site defines exosites in the catalytic domain.
Faseb J., 27, 2013
|
|
4FU4
 
 | | Human collagenase 3 (MMP-13) with peptide from pro-domain | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Stura, E.A, Vera, L, Visse, R, Nagase, H, Dive, V. | | Deposit date: | 2012-06-28 | | Release date: | 2013-08-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.849 Å) | | Cite: | Crystal structure of full-length human collagenase 3 (MMP-13) with peptides in the active site defines exosites in the catalytic domain.
Faseb J., 27, 2013
|
|
4EL4
 
 | | Crystal structure of the catalytic domain of botulinum neurotoxin BoNT/A C134S/C165S double mutant | | Descriptor: | 1,2-ETHANEDIOL, Botulinum neurotoxin A light chain, GLYCEROL, ... | | Authors: | Stura, E.A, Vera, L, Ptchelkine, D, Dive, V. | | Deposit date: | 2012-04-10 | | Release date: | 2012-08-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural Framework for Covalent Inhibition of Clostridium botulinum Neurotoxin A by Targeting Cys165.
J.Biol.Chem., 287, 2012
|
|
4EJ5
 
 | | Crystal structure of the catalytic domain of botulinum neurotoxin BoNT/A wild-type | | Descriptor: | 1,2-ETHANEDIOL, Botulinum neurotoxin A light chain, CARBONATE ION, ... | | Authors: | Stura, E.A, Vera, L, Dive, V. | | Deposit date: | 2012-04-06 | | Release date: | 2012-08-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural Framework for Covalent Inhibition of Clostridium botulinum Neurotoxin A by Targeting Cys165.
J.Biol.Chem., 287, 2012
|
|
5MG9
 
 | | Putative Ancestral Mamba toxin 1 (AncTx1-W28R/I38S) | | Descriptor: | 1,2-ETHANEDIOL, AncTx1-W28R/I38S, S-1,2-PROPANEDIOL, ... | | Authors: | Stura, E.A, Tepshi, L, Blanchet, G, Mourier, G, Servent, D. | | Deposit date: | 2016-11-21 | | Release date: | 2017-05-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Ancestral protein resurrection and engineering opportunities of the mamba aminergic toxins.
Sci Rep, 7, 2017
|
|
4WZV
 
 | | Crystal structure of a hydroxamate based inhibitor EN140 in complex with the MMP-9 catalytic domain | | Descriptor: | (2R)-4-(1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)-N-hydroxy-2-{[(4'-methoxybiphenyl-4-yl)sulfonyl](propan-2-yloxy)amino}butanamide, 1,2-ETHANEDIOL, AZIDE ION, ... | | Authors: | Stura, E.A, Vera, L, Cassar-Lajeunesse, E, Nuti, E, Dive, V, Rossello, A. | | Deposit date: | 2014-11-20 | | Release date: | 2015-08-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | N-O-Isopropyl Sulfonamido-Based Hydroxamates as Matrix Metalloproteinase Inhibitors: Hit Selection and in Vivo Antiangiogenic Activity.
J.Med.Chem., 58, 2015
|
|
4XCT
 
 | | Crystal structure of a hydroxamate based inhibitor ARP101 (EN73) in complex with the MMP-9 catalytic domain. | | Descriptor: | (2S,3S)-butane-2,3-diol, (2~{R})-3-methyl-~{N}-oxidanylidene-2-[(4-phenylphenyl)sulfonyl-propan-2-yloxy-amino]butanamide, 1,2-ETHANEDIOL, ... | | Authors: | Stura, E.A, Tepshi, L, Nuti, E, Dive, V, Cassar-Lajeunesse, E, Vera, L, Rossello, A. | | Deposit date: | 2014-12-18 | | Release date: | 2015-04-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | N-O-Isopropyl Sulfonamido-Based Hydroxamates as Matrix Metalloproteinase Inhibitors: Hit Selection and in Vivo Antiangiogenic Activity.
J.Med.Chem., 58, 2015
|
|
2C89
 
 | | Structure of the wild-type C3bot1 Exoenzyme (Free state, crystal form I) | | Descriptor: | MONO-ADP-RIBOSYLTRANSFERASE C3, SULFATE ION | | Authors: | Stura, E.A, Menetrey, J, Flatau, G, Boquet, P, Menez, A. | | Deposit date: | 2005-12-03 | | Release date: | 2007-02-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Basis for the Nad-Hydrolysis Mechanism and the Artt-Loop Plasticity of C3 Exoenzymes.
Protein Sci., 17, 2008
|
|
2C8C
 
 | | Structure of the ARTT motif Q212A mutant C3bot1 Exoenzyme (NAD-bound state, crystal form I) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MONO-ADP-RIBOSYLTRANSFERASE C3, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Stura, E.A, Menetrey, J, Flatau, G, Boquet, P, Menez, A. | | Deposit date: | 2005-12-03 | | Release date: | 2007-02-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis for the Nad-Hydrolysis Mechanism and the Artt-Loop Plasticity of C3 Exoenzymes.
Protein Sci., 17, 2008
|
|
2C8D
 
 | | Structure of the ARTT motif Q212A mutant C3bot1 Exoenzyme (Free state, crystal form I) | | Descriptor: | MONO-ADP-RIBOSYLTRANSFERASE C3, SULFATE ION | | Authors: | Stura, E.A, Menetrey, J, Flatau, G, Boquet, P, Menez, A. | | Deposit date: | 2005-12-03 | | Release date: | 2007-02-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for the Nad-Hydrolysis Mechanism and the Artt-Loop Plasticity of C3 Exoenzymes.
Protein Sci., 17, 2008
|
|
2C8E
 
 | | Structure of the ARTT motif E214N mutant C3bot1 Exoenzyme (Free state, crystal form III) | | Descriptor: | MONO-ADP-RIBOSYLTRANSFERASE C3, SULFATE ION | | Authors: | Stura, E.A, Menetrey, J, Flatau, G, Boquet, P, Menez, A. | | Deposit date: | 2005-12-03 | | Release date: | 2007-02-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Basis for the Nad-Hydrolysis Mechanism and the Artt-Loop Plasticity of C3 Exoenzymes.
Protein Sci., 17, 2008
|
|
2C8B
 
 | | Structure of the ARTT motif Q212A mutant C3bot1 Exoenzyme (Free state, crystal form II) | | Descriptor: | MONO-ADP-RIBOSYLTRANSFERASE C3, SULFATE ION | | Authors: | Stura, E.A, Menetrey, J, Flatau, G, Boquet, P, Menez, A. | | Deposit date: | 2005-12-03 | | Release date: | 2007-02-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis for the Nad-Hydrolysis Mechanism and the Artt-Loop Plasticity of C3 Exoenzymes.
Protein Sci., 17, 2008
|
|
2C8F
 
 | | Structure of the ARTT motif E214N mutant C3bot1 Exoenzyme (NAD-bound state, crystal form III) | | Descriptor: | MONO-ADP-RIBOSYLTRANSFERASE C3, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Stura, E.A, Menetrey, J, Flatau, G, Boquet, P, Menez, A. | | Deposit date: | 2005-12-03 | | Release date: | 2007-02-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for the Nad-Hydrolysis Mechanism and the Artt-Loop Plasticity of C3 Exoenzymes.
Protein Sci., 17, 2008
|
|
2C8A
 
 | | Structure of the wild-type C3bot1 Exoenzyme (Nicotinamide-bound state, crystal form I) | | Descriptor: | MONO-ADP-RIBOSYLTRANSFERASE C3, NICOTINAMIDE, SULFATE ION | | Authors: | Stura, E.A, Menetrey, J, Flatau, G, Boquet, P, Menez, A. | | Deposit date: | 2005-12-03 | | Release date: | 2007-02-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis for the Nad-Hydrolysis Mechanism and the Artt-Loop Plasticity of C3 Exoenzymes.
Protein Sci., 17, 2008
|
|
5I12
 
 | | Crystal structure of the catalytic domain of MMP-9 in complex with a selective sugar-conjugated arylsulfonamide carboxylate water-soluble inhibitor (DC27). | | Descriptor: | (2R)-2-[{(E)-2-[({(2R,3R,4R,5S,6R)-3-(acetylamino)-4,5-bis(acetyloxy)-6-[(acetyloxy)methyl]tetrahydro-2H-pyran-2-yl}carbamothioyl)amino]ethenyl}(biphenyl-4-ylsulfonyl)amino]-3-methylbutanoic acid, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Stura, E.A, Rosalia, L, Cuffaro, D, Tepshi, L, Ciccone, L, Rossello, A. | | Deposit date: | 2016-02-05 | | Release date: | 2016-07-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Sugar-Based Arylsulfonamide Carboxylates as Selective and Water-Soluble Matrix Metalloproteinase-12 Inhibitors.
Chemmedchem, 11, 2016
|
|
5I3M
 
 | | Crystal structure of the catalytic domain of MMP-12 in complex with a selective sugar-conjugated thiourea-linked carboxylate zinc-chelator water-soluble inhibitor (DC31). | | Descriptor: | (2S)-2-{[2-({[(2R,3R,4R,5S,6R)-3-(acetylamino)-4,5-dihydroxy-6-(hydroxymethyl)tetrahydro-2H-pyran-2-yl]carbamothioyl}amino)ethyl](biphenyl-4-ylsulfonyl)amino}-3-methylbutanoic acid (non-preferred name), 1,2-ETHANEDIOL, 1,4-DIETHYLENE DIOXIDE, ... | | Authors: | Stura, E.A, Rosalia, L, Cuffaro, D, Tepshi, L, Ciccone, L, Rossello, A. | | Deposit date: | 2016-02-10 | | Release date: | 2016-07-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Sugar-Based Arylsulfonamide Carboxylates as Selective and Water-Soluble Matrix Metalloproteinase-12 Inhibitors.
Chemmedchem, 11, 2016
|
|
5I2Z
 
 | | Crystal structure of the catalytic domain of MMP-12 in complex with a selective sugar-conjugated triazole-linked carboxylate chelating water-soluble inhibitor (DC24). | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, DIMETHYL SULFOXIDE, ... | | Authors: | Stura, E.A, Rosalia, L, Cuffaro, D, Tepshi, L, Ciccone, L, Rossello, A. | | Deposit date: | 2016-02-09 | | Release date: | 2016-07-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Sugar-Based Arylsulfonamide Carboxylates as Selective and Water-Soluble Matrix Metalloproteinase-12 Inhibitors.
Chemmedchem, 11, 2016
|
|
5I0L
 
 | | Crystal structure of the catalytic domain of MMP-12 in complex with a selective sugar-conjugated arylsulfonamide carboxylate water-soluble inhibitor (DC27). | | Descriptor: | (2R)-2-[{(E)-2-[({(2R,3R,4R,5S,6R)-3-(acetylamino)-4,5-bis(acetyloxy)-6-[(acetyloxy)methyl]tetrahydro-2H-pyran-2-yl}carbamothioyl)amino]ethenyl}(biphenyl-4-ylsulfonyl)amino]-3-methylbutanoic acid, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Stura, E.A, Rosalia, L, Cuffaro, D, Tepshi, L, Ciccone, L, Rossello, A. | | Deposit date: | 2016-02-04 | | Release date: | 2016-07-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Sugar-Based Arylsulfonamide Carboxylates as Selective and Water-Soluble Matrix Metalloproteinase-12 Inhibitors.
Chemmedchem, 11, 2016
|
|
5I4O
 
 | | Crystal structure of the catalytic domain of MMP-12 in complex with a selective sugar-conjugated triazole-linked carboxylate zinc-chelator water-soluble inhibitor (DC28). | | Descriptor: | CALCIUM ION, Macrophage metalloelastase, N-({1-[2-(acetylamino)-2-deoxy-beta-D-glucopyranosyl]-1H-1,2,3-triazol-4-yl}methyl)-N-[([1,1'-biphenyl]-4-yl)sulfonyl]-D-valine, ... | | Authors: | Stura, E.A, Rosalia, L, Cuffaro, D, Tepshi, L, Ciccone, L, Rossello, A. | | Deposit date: | 2016-02-12 | | Release date: | 2016-07-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Sugar-Based Arylsulfonamide Carboxylates as Selective and Water-Soluble Matrix Metalloproteinase-12 Inhibitors.
Chemmedchem, 11, 2016
|
|
4TQI
 
 | | Human transthyretin (TTR) complexed with 3-(9H-fluoren-9-ylideneaminooxy)propanoic acid in a dual binding mode | | Descriptor: | (2S)-3-[(9H-fluoren-9-ylideneamino)oxy]-2-methylpropanoic acid, GLYCEROL, Transthyretin | | Authors: | Stura, E.A, Ciccone, L, Nencetti, S, Rossello, A, Orlandini, E. | | Deposit date: | 2014-06-11 | | Release date: | 2015-06-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | X-ray crystal structure and activity of fluorenyl-based compounds as transthyretin fibrillogenesis inhibitors.
J Enzyme Inhib Med Chem, 2015
|
|
