1SPY
 
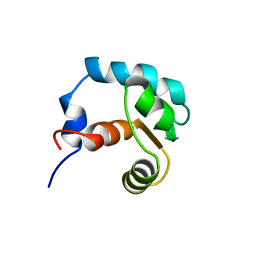 | | REGULATORY DOMAIN OF HUMAN CARDIAC TROPONIN C IN THE CALCIUM-FREE STATE, NMR, 40 STRUCTURES | | Descriptor: | TROPONIN C | | Authors: | Spyracopoulos, L, Li, M.X, Sia, S.K, Gagne, S.M, Chandra, M, Solaro, R.J, Sykes, B.D. | | Deposit date: | 1997-07-14 | | Release date: | 1998-09-16 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Calcium-induced structural transition in the regulatory domain of human cardiac troponin C.
Biochemistry, 36, 1997
|
|
1AP4
 
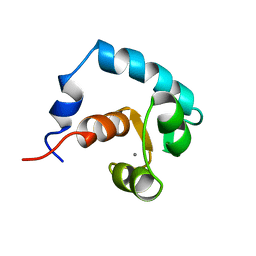 | | REGULATORY DOMAIN OF HUMAN CARDIAC TROPONIN C IN THE CALCIUM-SATURATED STATE, NMR, 40 STRUCTURES | | Descriptor: | CALCIUM ION, CARDIAC N-TROPONIN C | | Authors: | Li, M.X, Spyracopoulos, L, Sia, S.K, Gagne, S.M, Chandra, M, Solaro, R.J, Sykes, B.D. | | Deposit date: | 1997-07-24 | | Release date: | 1998-07-15 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Calcium-induced structural transition in the regulatory domain of human cardiac troponin C.
Biochemistry, 36, 1997
|
|
1AJ4
 
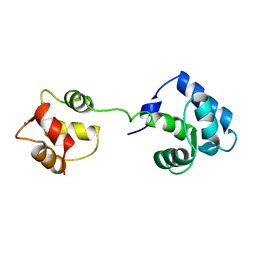 | | STRUCTURE OF CALCIUM-SATURATED CARDIAC TROPONIN C, NMR, 1 STRUCTURE | | Descriptor: | CALCIUM ION, TROPONIN C | | Authors: | Sia, S.K, Li, M.X, Spyracopoulos, L, Gagne, S.M, Liu, W, Putkey, J.A, Sykes, B.D. | | Deposit date: | 1997-05-14 | | Release date: | 1998-05-20 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | Structure of cardiac muscle troponin C unexpectedly reveals a closed regulatory domain.
J.Biol.Chem., 272, 1997
|
|
1ZAC
 
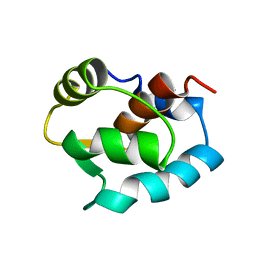 | | N-DOMAIN OF TROPONIN C FROM CHICKEN SKELETAL MUSCLE, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | TROPONIN-C | | Authors: | Tsuda, S, Miura, A, Gagne, S.M, Spyracopoulos, L, Sykes, B.D. | | Deposit date: | 1998-04-07 | | Release date: | 1998-11-11 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Low-temperature-induced structural changes in the Apo regulatory domain of skeletal muscle troponin C.
Biochemistry, 38, 1999
|
|
3CTN
 
 | | STRUCTURE OF CALCIUM-SATURATED CARDIAC TROPONIN C, NMR, 30 STRUCTURES | | Descriptor: | CALCIUM ION, TROPONIN C | | Authors: | Sia, S.K, Li, M.X, Spyracopoulos, L, Gagne, S.M, Liu, W, Putkey, J.A, Sykes, B.D. | | Deposit date: | 1997-05-08 | | Release date: | 1998-05-13 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Structure of cardiac muscle troponin C unexpectedly reveals a closed regulatory domain.
J.Biol.Chem., 272, 1997
|
|
1SKT
 
 | | SOLUTION STRUCTURE OF APO N-DOMAIN OF TROPONIN C, NMR, 40 STRUCTURES | | Descriptor: | TROPONIN-C | | Authors: | Tsuda, S, Miura, A, Gagne, S.M, Spyracopoulos, L, Sykes, B.D. | | Deposit date: | 1998-04-06 | | Release date: | 1998-12-09 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Low-temperature-induced structural changes in the Apo regulatory domain of skeletal muscle troponin C.
Biochemistry, 38, 1999
|
|
1JC2
 
 | | COMPLEX OF THE C-DOMAIN OF TROPONIN C WITH RESIDUES 1-40 OF TROPONIN I | | Descriptor: | CALCIUM ION, TROPONIN C, SKELETAL MUSCLE | | Authors: | Mercier, P, Spyracopoulos, L, Sykes, B.D. | | Deposit date: | 2001-06-07 | | Release date: | 2001-09-14 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Structure, dynamics, and thermodynamics of the structural domain of troponin C in complex with the regulatory peptide 1-40 of troponin I
Biochemistry, 40, 2001
|
|
1MXL
 
 | |
1ZGU
 
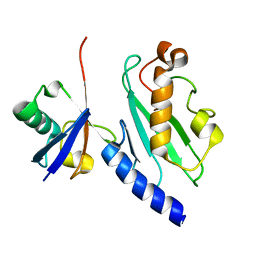 | | Solution structure of the human Mms2-Ubiquitin complex | | Descriptor: | Ubiquitin, Ubiquitin-conjugating enzyme E2 variant 2 | | Authors: | Lewis, M.J, Saltibus, L.F, Hau, D.D, Xiao, W, Spyracopoulos, L. | | Deposit date: | 2005-04-22 | | Release date: | 2006-04-04 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Non-Covalent Interaction Between Ubiquitin and the Ubiquitin Conjugating Enzyme Variant Human MMS2.
J.Biomol.Nmr, 34, 2006
|
|
2HLW
 
 | | Solution Structure of the Human Ubiquitin-conjugating Enzyme Variant Uev1a | | Descriptor: | Ubiquitin-conjugating enzyme E2 variant 1 | | Authors: | Hau, D.D, Lewis, M.J, Saltibus, L.F, Pastushok, L, Xiao, W, Spyracopoulos, L. | | Deposit date: | 2006-07-10 | | Release date: | 2006-09-05 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Structure and interactions of the ubiquitin-conjugating enzyme variant human uev1a: implications for enzymatic synthesis of polyubiquitin chains(,).
Biochemistry, 45, 2006
|
|
2CTN
 
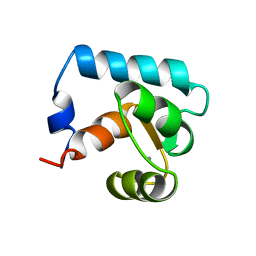 | | STRUCTURE OF CALCIUM-SATURATED CARDIAC TROPONIN C, NMR, 30 STRUCTURES | | Descriptor: | CALCIUM ION, TROPONIN C | | Authors: | Sia, S.K, Li, M.X, Spyracopoulos, L, Gagne, S.M, Liu, W, Putkey, J.A, Sykes, B.D. | | Deposit date: | 1997-05-06 | | Release date: | 1998-05-06 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | Structure of cardiac muscle troponin C unexpectedly reveals a closed regulatory domain.
J.Biol.Chem., 272, 1997
|
|
1IH0
 
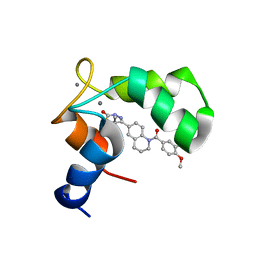 | | Structure of the C-domain of Human Cardiac Troponin C in Complex with Ca2+ Sensitizer EMD 57033 | | Descriptor: | 5-[1-(3,4-DIMETHOXY-BENZOYL)-1,2,3,4-TETRAHYDRO-QUINOLIN-6-YL]-6-METHYL-3,6-DIHYDRO-[1,3,4]THIADIAZIN-2-ONE, CALCIUM ION, TROPONIN C, ... | | Authors: | Wang, X, Li, M.X, Spyracopoulos, L, Beier, N, Chandra, M, Solaro, R.J, Sykes, B.D. | | Deposit date: | 2001-04-18 | | Release date: | 2001-10-10 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Structure of the C-domain of human cardiac troponin C in complex with the Ca2+ sensitizing drug EMD 57033.
J.Biol.Chem., 276, 2001
|
|
1L1I
 
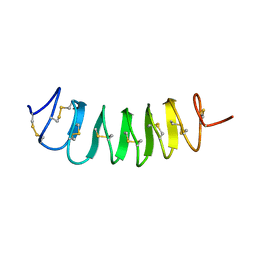 | | Solution Structure of the Tenebrio molitor Antifreeze Protein | | Descriptor: | Thermal hysteresis protein isoform YL-1 (2-14) | | Authors: | Daley, M.E, Spyracopoulos, L, Jia, Z, Davies, P.L, Sykes, B.D. | | Deposit date: | 2002-02-16 | | Release date: | 2002-05-22 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Structure and dynamics of a beta-helical antifreeze protein.
Biochemistry, 41, 2002
|
|
2MKF
 
 | |
2MKG
 
 | |
2N9E
 
 | |
1N4I
 
 | | Solution structure of spruce budworm antifreeze protein at 5 degrees celsius | | Descriptor: | thermal hysteresis protein | | Authors: | Graether, S.P, Gagne, S.M, Spyracopoulos, L, Jia, Z, Davies, P.L, Sykes, B.D. | | Deposit date: | 2002-10-31 | | Release date: | 2003-04-08 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Spruce Budworm Antifreeze Protein: Changes in Structure and Dynamics at Low Temperature
J.Mol.Biol., 327, 2003
|
|
1RD4
 
 | | An allosteric inhibitor of LFA-1 bound to its I-domain | | Descriptor: | 1-ACETYL-4-(4-{4-[(2-ETHOXYPHENYL)THIO]-3-NITROPHENYL}PYRIDIN-2-YL)PIPERAZINE, Integrin alpha-L | | Authors: | Crump, M.P, Ceska, T.A, Spyracopoulos, L, Henry, A, Archibald, S.C, Alexander, R, Taylor, R.J, Findlow, S.C, O'Connell, J, Robinson, M.K, Shock, A. | | Deposit date: | 2003-11-05 | | Release date: | 2004-03-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of an allosteric inhibitor of LFA-1 bound to the I-domain studied by crystallography, NMR, and calorimetry
Biochemistry, 43, 2004
|
|
2JMD
 
 | | Solution Structure of the Ring Domain of Human TRAF6 | | Descriptor: | TNF receptor-associated factor 6, ZINC ION | | Authors: | Mercier, P, Lewis, M.J, Hau, D.D, Saltibus, L.F, Xiao, W, Spyracopoulos, L. | | Deposit date: | 2006-11-02 | | Release date: | 2007-04-10 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Structure, interactions, and dynamics of the RING domain from human TRAF6
Protein Sci., 16, 2007
|
|
1C8A
 
 | |
1C89
 
 | |
3LRU
 
 | |
4MA7
 
 | | Crystal structure of mouse prion protein complexed with Promazine | | Descriptor: | Major prion protein, POM1 heavy chain, POM1 light chain, ... | | Authors: | Baral, P.K, Swayampakula, M, James, M.N.G. | | Deposit date: | 2013-08-15 | | Release date: | 2014-01-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural basis of prion inhibition by phenothiazine compounds.
Structure, 22, 2014
|
|
4MA8
 
 | | Crystal structure of mouse prion protein complexed with Chlorpromazine | | Descriptor: | 3-(2-chloro-10H-phenothiazin-10-yl)-N,N-dimethylpropan-1-amine, Major prion protein, POM1 heavy chain, ... | | Authors: | Baral, P.K, Swayampakula, M, James, M.N.G. | | Deposit date: | 2013-08-15 | | Release date: | 2014-01-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of prion inhibition by phenothiazine compounds.
Structure, 22, 2014
|
|
4NR3
 
 | |
