1BX2
 
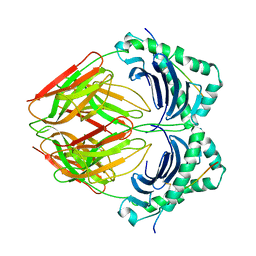 | | CRYSTAL STRUCTURE OF HLA-DR2 (DRA*0101,DRB1*1501) COMPLEXED WITH A PEPTIDE FROM HUMAN MYELIN BASIC PROTEIN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PROTEIN (HLA-DR2) | | Authors: | Smith, K.J, Pyrdol, J, Gauthier, L, Wiley, D.C, Wucherpfennig, K. | | Deposit date: | 1998-10-12 | | Release date: | 1998-10-21 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of HLA-DR2 (DRA*0101, DRB1*1501) complexed with a peptide from human myelin basic protein.
J.Exp.Med., 188, 1998
|
|
1A1O
 
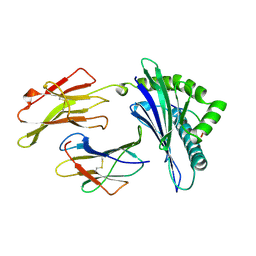 | | MHC CLASS I MOLECULE B*5301 COMPLEXED WITH PEPTIDE LS6 (KPIVQYDNF) FROM THE MALARIA PARASITE P. FALCIPARUM | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, BW-53 B*5301 alpha chain, ... | | Authors: | Smith, K.J, Reid, S.W, Harlos, K, Mcmichael, A.J, Stuart, D.I, Bell, J.I, Jones, E.Y. | | Deposit date: | 1997-12-11 | | Release date: | 1998-04-08 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Bound water structure and polymorphic amino acids act together to allow the binding of different peptides to MHC class I HLA-B53.
Immunity, 4, 1996
|
|
1A1N
 
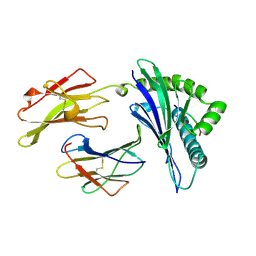 | | MHC CLASS I MOLECULE B*3501 COMPLEXED WITH PEPTIDE VPLRPMTY FROM THE NEF PROTEIN (75-82) OF HIV1 | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, BW-53 B*5301 alpha chain, ... | | Authors: | Smith, K.J, Reid, S.W, Stuart, D.I, Mcmichael, A.J, Jones, E.Y, Bell, J.I. | | Deposit date: | 1997-12-11 | | Release date: | 1998-04-08 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | An altered position of the alpha 2 helix of MHC class I is revealed by the crystal structure of HLA-B*3501.
Immunity, 4, 1996
|
|
1A1M
 
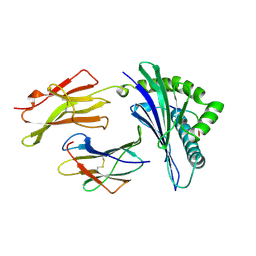 | | MHC CLASS I MOLECULE B*5301 COMPLEXED WITH PEPTIDE TPYDINQML FROM GAG PROTEIN OF HIV2 | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, BW-53 B*5301 alpha chain, ... | | Authors: | Smith, K.J, Reid, S.W, Harlos, K, Mcmichael, A.J, Stuart, D.I, Bell, J.I, Jones, E.Y. | | Deposit date: | 1997-12-11 | | Release date: | 1998-04-08 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Bound water structure and polymorphic amino acids act together to allow the binding of different peptides to MHC class I HLA-B53.
Immunity, 4, 1996
|
|
1VZO
 
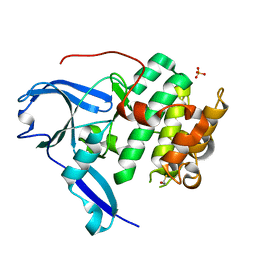 | | The structure of the N-terminal kinase domain of MSK1 reveals a novel autoinhibitory conformation for a dual kinase protein | | Descriptor: | BETA-MERCAPTOETHANOL, RIBOSOMAL PROTEIN S6 KINASE ALPHA 5, SULFATE ION | | Authors: | Smith, K.J, Carter, P.S, Bridges, A, Horrocks, P, Lewis, C, Pettman, G, Clarke, A, Brown, M, Hughes, J, Wilkinson, M, Bax, B, Reith, A. | | Deposit date: | 2004-05-21 | | Release date: | 2004-06-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Structure of Msk1 Reveals a Novel Autoinhibitory Conformation for a Dual Kinase Protein
Structure, 12, 2004
|
|
2AIA
 
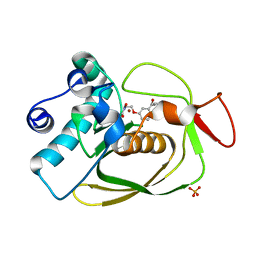 | | S.pneumoniae PDF complexed with SB-543668 | | Descriptor: | 2-(3-BENZOYLPHENOXY)ETHYL(HYDROXY)FORMAMIDE, NICKEL (II) ION, Peptide deformylase, ... | | Authors: | Smith, K.J, Petit, C.M, Aubart, K, Smyth, M, McManus, E, Jones, J, Fosberry, A, Lewis, C, Lonetto, M, Christensen, S.B. | | Deposit date: | 2005-07-29 | | Release date: | 2005-09-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Variation and inhibitor binding in polypeptide deformylase from four different bacterial species.
Protein Sci., 12, 2003
|
|
2AI9
 
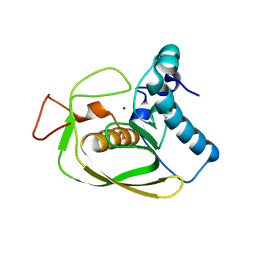 | | S.aureus Polypeptide Deformylase | | Descriptor: | NICKEL (II) ION, Peptide deformylase, SULFATE ION | | Authors: | Smith, K.J, Petit, C.M, Aubart, K, Smyth, M, McManus, E, Jones, J, Fosberry, A, Lewis, C, Lonetto, M, Christensen, S.B. | | Deposit date: | 2005-07-29 | | Release date: | 2005-09-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Variation and inhibitor binding in polypeptide deformylase from four different bacterial species.
Protein Sci., 12, 2003
|
|
2AIE
 
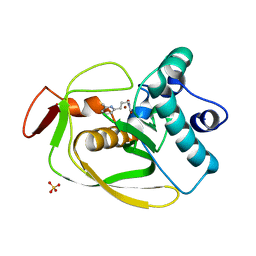 | | S.pneumoniae polypeptide deformylase complexed with SB-505684 | | Descriptor: | HYDROXY[3-(6-METHYLPYRIDIN-2-YL)PROPYL]FORMAMIDE, NICKEL (II) ION, Peptide deformylase, ... | | Authors: | Smith, K.J, Petit, C.M, Aubart, K, Smyth, M, McManus, E, Jones, J, Fosberry, A, Lewis, C, Lonetto, M, Christensen, S.B. | | Deposit date: | 2005-07-29 | | Release date: | 2005-09-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Variation and inhibitor binding in polypeptide deformylase from four different bacterial species
Protein Sci., 12, 2003
|
|
2AI7
 
 | | S.pneumoniae Polypeptide Deformylase complexed with SB-485345 | | Descriptor: | NICKEL (II) ION, Peptide deformylase, SULFATE ION, ... | | Authors: | Smith, K.J, Petit, C.M, Aubart, K, Smyth, M, McManus, E, Jones, J, Fosberry, A, Lewis, C, Lonetto, M, Christensen, S.B. | | Deposit date: | 2005-07-29 | | Release date: | 2005-09-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Variation and inhibitor binding in polypeptide deformylase from four different bacterial species
Protein Sci., 12, 2003
|
|
2AI8
 
 | | E.coli Polypeptide Deformylase complexed with SB-485343 | | Descriptor: | NICKEL (II) ION, Peptide deformylase, [HYDROXY(3-PHENYLPROPYL)AMINO]METHANOL | | Authors: | Smith, K.J, Petit, C.M, Aubart, K, Smyth, M, McManus, E, Jones, J, Fosberry, A, Lewis, C, Lonetto, M, Christensen, S.B. | | Deposit date: | 2005-07-29 | | Release date: | 2005-09-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Variation and inhibitor binding in polypeptide deformylase from four different bacterial species.
Protein Sci., 12, 2003
|
|
1LOI
 
 | | N-TERMINAL SPLICE REGION OF RAT C-AMP PHOSPHODIESTERASE, NMR, 7 STRUCTURES | | Descriptor: | CYCLIC 3',5'-AMP SPECIFIC PHOSPHODIESTERASE RD1 | | Authors: | Smith, K.J, Scotland, G, Beattie, J, Trayer, I.P, Houslay, M.D. | | Deposit date: | 1996-05-21 | | Release date: | 1997-05-15 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Determination of the structure of the N-terminal splice region of the cyclic AMP-specific phosphodiesterase RD1 (RNPDE4A1) by 1H NMR and identification of the membrane association domain using chimeric constructs.
J.Biol.Chem., 271, 1996
|
|
1E9K
 
 | | The structure of the RACK1 interaction sites located within the unique N-terminal region of the cAMP-specific phosphodiesterase, PDE4D5. | | Descriptor: | cAMP-specific 3',5'-cyclic phosphodiesterase 4D | | Authors: | Bolger, G.B, Smith, K.J, McCahill, A, Hyde, E.I, Steele, M.R, Houslay, M.D. | | Deposit date: | 2000-10-20 | | Release date: | 2001-10-18 | | Last modified: | 2018-06-20 | | Method: | SOLUTION NMR | | Cite: | 1H NMR structural and functional characterisation of a cAMP-specific phosphodiesterase-4D5 (PDE4D5) N-terminal region peptide that disrupts PDE4D5 interaction with the signalling scaffold proteins, beta-arrestin and RACK1.
Cell. Signal., 19, 2007
|
|
1AGB
 
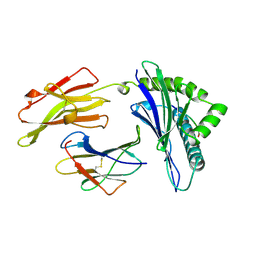 | | ANTAGONIST HIV-1 GAG PEPTIDES INDUCE STRUCTURAL CHANGES IN HLA B8-HIV-1 GAG PEPTIDE (GGRKKYKL-3R MUTATION) | | Descriptor: | B*0801, BETA-2 MICROGLOBULIN, HIV-1 GAG PEPTIDE (GGRKKYKL - 3R MUTATION) | | Authors: | Reid, S.W, Mcadam, S, Smith, K.J, Klenerman, P, O'Callaghan, C.A, Harlos, K, Jakobsen, B.K, Mcmichael, A.J, Bell, J, Stuart, D.I, Jones, E.Y. | | Deposit date: | 1997-03-24 | | Release date: | 1997-06-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Antagonist HIV-1 Gag peptides induce structural changes in HLA B8.
J.Exp.Med., 184, 1996
|
|
1AGC
 
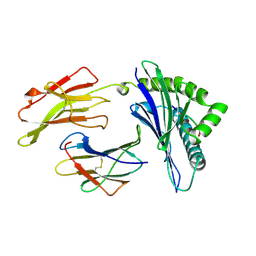 | | ANTAGONIST HIV-1 GAG PEPTIDES INDUCE STRUCTURAL CHANGES IN HLA B8-HIV-1 GAG PEPTIDE (GGKKKYQL-7Q MUTATION) | | Descriptor: | B*0801, BETA-2 MICROGLOBULIN, HIV-1 GAG PEPTIDE (GGKKKYQL - 7Q MUTATION) | | Authors: | Reid, S.W, Mcadam, S, Smith, K.J, Klenerman, P, O'Callaghan, C.A, Harlos, K, Jakobsen, B.K, Mcmichael, A.J, Bell, J, Stuart, D.I, Jones, E.Y. | | Deposit date: | 1997-03-24 | | Release date: | 1997-06-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Antagonist HIV-1 Gag peptides induce structural changes in HLA B8.
J.Exp.Med., 184, 1996
|
|
1AGF
 
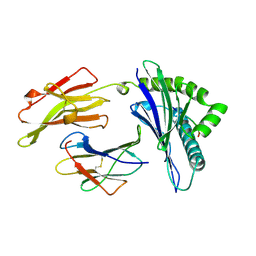 | | ANTAGONIST HIV-1 GAG PEPTIDES INDUCE STRUCTURAL CHANGES IN HLA B8-HIV-1 GAG PEPTIDE (GGKKRYKL-5R MUTATION) | | Descriptor: | B*0801, BETA-2 MICROGLOBULIN, HIV-1 GAG PEPTIDE (GGKKRYKL - 5R MUTATION) | | Authors: | Reid, S.W, Mcadam, S, Smith, K.J, Klenerman, P, O'Callaghan, C.A, Harlos, K, Jakobsen, B.K, Mcmichael, A.J, Bell, J, Stuart, D.I, Jones, E.Y. | | Deposit date: | 1997-03-24 | | Release date: | 1997-06-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Antagonist HIV-1 Gag peptides induce structural changes in HLA B8.
J.Exp.Med., 184, 1996
|
|
1AGE
 
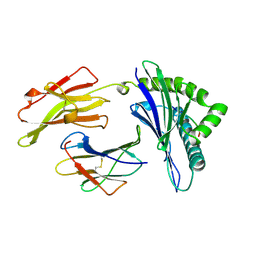 | | ANTAGONIST HIV-1 GAG PEPTIDES INDUCE STRUCTURAL CHANGES IN HLA B8-HIV-1 GAG PEPTIDE (GGKKKYRL-7R MUTATION) | | Descriptor: | B*0801, BETA-2 MICROGLOBULIN, HIV-1 GAG PEPTIDE (GGKKKYRL - 7R MUTATION) | | Authors: | Reid, S.W, Mcadam, S, Smith, K.J, Klenerman, P, O'Callaghan, C.A, Harlos, K, Jakobsen, B.K, Mcmichael, A.J, Bell, J, Stuart, D.I, Jones, E.Y. | | Deposit date: | 1997-03-24 | | Release date: | 1997-06-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Antagonist HIV-1 Gag peptides induce structural changes in HLA B8.
J.Exp.Med., 184, 1996
|
|
1AGD
 
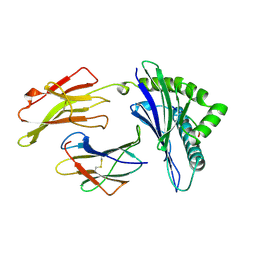 | | ANTAGONIST HIV-1 GAG PEPTIDES INDUCE STRUCTURAL CHANGES IN HLA B8-HIV-1 GAG PEPTIDE (GGKKKYKL-INDEX PEPTIDE) | | Descriptor: | B*0801, BETA-2 MICROGLOBULIN, HIV-1 GAG PEPTIDE (GGKKKYKL - INDEX PEPTIDE) | | Authors: | Reid, S.W, Mcadam, S, Smith, K.J, Klenerman, P, O'Callaghan, C.A, Harlos, K, Jakobsen, B.K, Mcmichael, A.J, Bell, J, Stuart, D.I, Jones, E.Y. | | Deposit date: | 1997-03-24 | | Release date: | 1997-06-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Antagonist HIV-1 Gag peptides induce structural changes in HLA B8.
J.Exp.Med., 184, 1996
|
|
1B0R
 
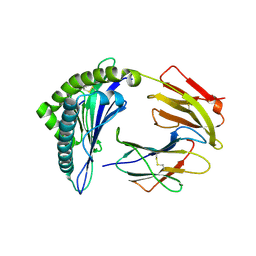 | | CRYSTAL STRUCTURE OF HLA-A*0201 COMPLEXED WITH A PEPTIDE WITH THE CARBOXYL-TERMINAL GROUP SUBSTITUTED BY A METHYL GROUP | | Descriptor: | PROTEIN (HLA-A*0201), PROTEIN (INFLUENZA MATRIX PEPTIDE) | | Authors: | Bouvier, M, Guo, H, Smith, K.J, Wiley, D.C. | | Deposit date: | 1998-11-12 | | Release date: | 1999-11-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structures of HLA-A*0201 complexed with antigenic peptides with either the amino- or carboxyl-terminal group substituted by a methyl group.
Proteins, 33, 1998
|
|
1BD2
 
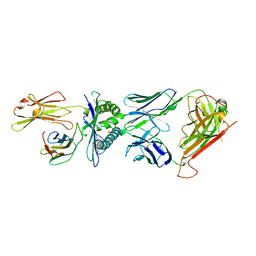 | | COMPLEX BETWEEN HUMAN T-CELL RECEPTOR B7, VIRAL PEPTIDE (TAX) AND MHC CLASS I MOLECULE HLA-A 0201 | | Descriptor: | BETA-2 MICROGLOBULIN, HLA-A 0201, T CELL RECEPTOR ALPHA, ... | | Authors: | Ding, Y.-H, Smith, K.J, Garboczi, D.N, Utz, U, Biddison, W.E, Wiley, D.C. | | Deposit date: | 1998-05-12 | | Release date: | 1998-08-19 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Two human T cell receptors bind in a similar diagonal mode to the HLA-A2/Tax peptide complex using different TCR amino acids.
Immunity, 8, 1998
|
|
3SW8
 
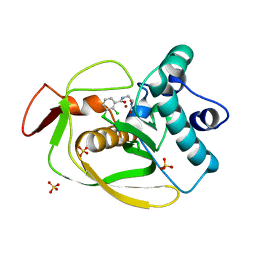 | |
6NZN
 
 | | Dimer-of-dimer amyloid fibril structure of glucagon | | Descriptor: | Glucagon | | Authors: | Gelenter, M.D, Smith, K.J, Liao, S.Y, Mandala, V.S, Dregni, A.J, Lamm, M.S, Tian, Y, Wei, X, Pochan, D.J, Tucker, T.J, Su, Y, Hong, M. | | Deposit date: | 2019-02-14 | | Release date: | 2019-06-05 | | Last modified: | 2023-06-14 | | Method: | SOLID-STATE NMR | | Cite: | The peptide hormone glucagon forms amyloid fibrils with two coexisting beta-strand conformations.
Nat.Struct.Mol.Biol., 26, 2019
|
|
2XHD
 
 | | Crystal structure of N-((2S)-5-(6-fluoro-3-pyridinyl)-2,3-dihydro-1H- inden-2-yl)-2-propanesulfonamide in complex with the ligand binding domain of the human GluA2 receptor | | Descriptor: | GLUTAMATE RECEPTOR 2, GLUTAMIC ACID, N-[(2S)-5-(6-FLUORO-3-PYRIDINYL)-2,3-DIHYDRO-1H-INDEN-2-YL]-2-PROPANESULFONAMIDE, ... | | Authors: | Ward, S.E, Harries, M, Aldegheri, L, Andreotti, D, Ballantine, S, Bax, B.D, Harris, A.J, Harker, A.J, Lund, J, Melarange, R, Mingardi, A, Mookherjee, C, Mosley, J, Neve, M, Oliosi, B, Profeta, R, Smith, K.J, Smith, P.W, Spada, S, Thewlis, K.M, Yusaf, S.P. | | Deposit date: | 2010-06-14 | | Release date: | 2010-07-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of N-[(2S)-5-(6-Fluoro-3-Pyridinyl)-2,3-Dihydro-1H-Inden-2-Yl]-2-Propanesulfonamide, a Novel Clinical Ampa Receptor Positive Modulator.
J.Med.Chem., 53, 2010
|
|
4BHN
 
 | | Crystal Structures of Ask1-inhibitor Complexes | | Descriptor: | 4-tert-butyl-N-[6-(1H-pyrazol-4-yl)imidazo[1,2-a]pyridin-2-yl]benzamide, GLYCEROL, MITOGEN-ACTIVATED PROTEIN KINASE KINASE KINASE 5 | | Authors: | Singh, O, Shillings, A, Craggs, P, Wall, I, Rowland, P, Skarzynski, T, Hobbs, C.I, Hardwick, P, Tanner, R, Blunt, M, Witty, D.R, Smith, K.J. | | Deposit date: | 2013-04-04 | | Release date: | 2013-07-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of Ask1-Inhibtor Complexes Provide a Platform for Structure Based Drug Design.
Protein Sci., 22, 2013
|
|
4BIC
 
 | | Crystal Structures of Ask1-inhibitor Complexes | | Descriptor: | GLYCEROL, MITOGEN-ACTIVATED PROTEIN KINASE KINASE KINASE 5, N-(2-aminoethyl)-5-{1H-pyrrolo[2,3-b]pyridin-3-yl}thiophene-2-sulfonamide | | Authors: | Singh, O, Shillings, A, Craggs, P, Wall, I, Rowland, P, Skarzynski, T, Hobbs, C.I, Hardwick, P, Tanner, R, Blunt, M, Witty, D.R, Smith, K.J. | | Deposit date: | 2013-04-10 | | Release date: | 2013-07-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Crystal Structures of Ask1-Inhibtor Complexes Provide a Platform for Structure Based Drug Design.
Protein Sci., 22, 2013
|
|
4BIE
 
 | | Crystal Structures of Ask1-inhibitor Complexes | | Descriptor: | GLYCEROL, MITOGEN-ACTIVATED PROTEIN KINASE KINASE KINASE 5, N-(2-aminoethyl)-5-{2-methyl-1H-pyrrolo[2,3-b]pyridin-4-yl}thiophene-2-sulfonamide | | Authors: | Singh, O, Shillings, A, Craggs, P, Wall, I, Rowland, P, Skarzynski, T, Hobbs, C.I, Hardwick, P, Tanner, R, Blunt, M, Witty, D.R, Smith, K.J. | | Deposit date: | 2013-04-10 | | Release date: | 2013-07-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Crystal Structures of Ask1-Inhibtor Complexes Provide a Platform for Structure Based Drug Design.
Protein Sci., 22, 2013
|
|
