5X19
 
 | | CO bound cytochrome c oxidase at 100 micro sec after pump laser irradiation to release CO from O2 reduction center | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Shimada, A, Kubo, M, Baba, S, Yamashita, K, Hirata, K, Ueno, G, Nomura, T, Kimura, T, Shinzawa-Itoh, K, Baba, J, Hatano, K, Eto, Y, Miyamoto, A, Murakami, H, Kumasaka, T, Owada, S, Tono, K, Yabashi, M, Yamaguchi, Y, Yanagisawa, S, Sakaguchi, M, Ogura, T, Komiya, R, Yan, J, Yamashita, E, Yamamoto, M, Ago, H, Yoshikawa, S, Tsukihara, T. | | Deposit date: | 2017-01-25 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A nanosecond time-resolved XFEL analysis of structural changes associated with CO release from cytochrome c oxidase.
Sci Adv, 3, 2017
|
|
5X1B
 
 | | CO bound cytochrome c oxidase at 20 nsec after pump laser irradiation to release CO from O2 reduction center | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Shimada, A, Kubo, M, Baba, S, Yamashita, K, Hirata, K, Ueno, G, Nomura, T, Kimura, T, Shinzawa-Itoh, K, Baba, J, Hatano, K, Eto, Y, Miyamoto, A, Murakami, H, Kumasaka, T, Owada, S, Tono, K, Yabashi, M, Yamaguchi, Y, Yanagisawa, S, Sakaguchi, M, Ogura, T, Komiya, R, Yan, J, Yamashita, E, Yamamoto, M, Ago, H, Yoshikawa, S, Tsukihara, T. | | Deposit date: | 2017-01-25 | | Release date: | 2017-08-09 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A nanosecond time-resolved XFEL analysis of structural changes associated with CO release from cytochrome c oxidase.
Sci Adv, 3, 2017
|
|
5X1F
 
 | | CO bound cytochrome c oxidase without pump laser irradiation at 278K | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Shimada, A, Kubo, M, Baba, S, Yamashita, K, Hirata, K, Ueno, G, Nomura, T, Kimura, T, Shinzawa-Itoh, K, Baba, J, Hatano, K, Eto, Y, Miyamoto, A, Murakami, H, Kumasaka, T, Owada, S, Tono, K, Yabashi, M, Yamaguchi, Y, Yanagisawa, S, Sakaguchi, M, Ogura, T, Komiya, R, Yan, J, Yamashita, E, Yamamoto, M, Ago, H, Yoshikawa, S, Tsukihara, T. | | Deposit date: | 2017-01-25 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A nanosecond time-resolved XFEL analysis of structural changes associated with CO release from cytochrome c oxidase.
Sci Adv, 3, 2017
|
|
7VVR
 
 | | Bovine cytochrome c oxidese in CN-bound mixed valence state at 50 K | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Shimada, A, Tsukihara, T. | | Deposit date: | 2021-11-08 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystallographic cyanide-probing for cytochrome c oxidase reveals structural bases suggesting that a putative proton transfer H-pathway pumps protons.
J.Biol.Chem., 299, 2023
|
|
7VUW
 
 | | Bovine heart cytochrome c oxidase in the cyanide-bound fully oxidized state at 50 K | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Shimada, A, Tsukihara, T. | | Deposit date: | 2021-11-04 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystallographic cyanide-probing for cytochrome c oxidase reveals structural bases suggesting that a putative proton transfer H-pathway pumps protons.
J.Biol.Chem., 299, 2023
|
|
3ED1
 
 | | Crystal Structure of Rice GID1 complexed with GA3 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, GIBBERELLIN A3, Gibberellin receptor GID1, ... | | Authors: | Shimada, A, Nakatsu, T, Ueguchi-Tanaka, M, Kato, H, Matsuoka, M. | | Deposit date: | 2008-09-02 | | Release date: | 2008-11-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for gibberellin recognition by its receptor GID1.
Nature, 456, 2008
|
|
3EBL
 
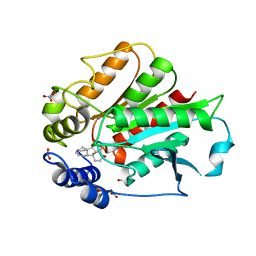 | | Crystal Structure of Rice GID1 complexed with GA4 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, GIBBERELLIN A4, Gibberellin receptor GID1, ... | | Authors: | Shimada, A, Nakatsu, T, Ueguchi-Tanaka, M, Kato, H, Matsuoka, M. | | Deposit date: | 2008-08-28 | | Release date: | 2008-11-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for gibberellin recognition by its receptor GID1.
Nature, 456, 2008
|
|
2CXY
 
 | |
1V9D
 
 | | Crystal structure of the core FH2 domain of mouse mDia1 | | Descriptor: | Diaphanous protein homolog 1, SULFATE ION | | Authors: | Shimada, A, Nyitrai, M, Vetter, I.R, Kuhlmann, D, Bugyi, B, Narumiya, S, Geeves, M.A, Wittinghofer, A. | | Deposit date: | 2004-01-24 | | Release date: | 2004-03-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The core FH2 domain of diaphanous-related formins is an elongated actin binding protein that inhibits polymerization.
Mol.Cell, 13, 2004
|
|
5AWR
 
 | |
5AWU
 
 | |
5AWS
 
 | |
5AWT
 
 | |
1IQ0
 
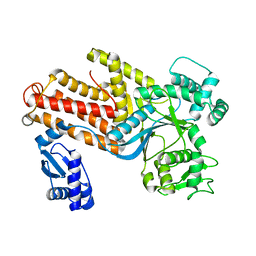 | | THERMUS THERMOPHILUS ARGINYL-TRNA SYNTHETASE | | Descriptor: | ARGINYL-TRNA SYNTHETASE | | Authors: | Shimada, A, Nureki, O, Goto, M, Takahashi, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2001-05-24 | | Release date: | 2001-11-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and mutational studies of the recognition of the arginine tRNA-specific major identity element, A20, by arginyl-tRNA synthetase.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
5ZCO
 
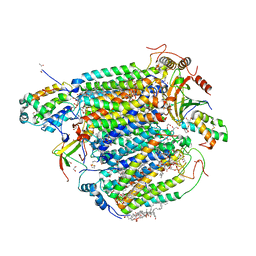 | | azide-bound cytochrome c oxidase structure determined using the crystals exposed to 2 mM azide solution for 2 days | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Shimada, A, Hatano, K, Tadehara, H, Tsukihara, T. | | Deposit date: | 2018-02-19 | | Release date: | 2018-08-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray structural analyses of azide-bound cytochromecoxidases reveal that the H-pathway is critically important for the proton-pumping activity.
J. Biol. Chem., 293, 2018
|
|
5B3S
 
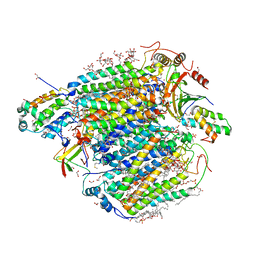 | | Bovine heart cytochrome c oxidase in the carbon monoxide-bound mixed-valence state at 1.68 angstrom resolution (50 K) | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Shimada, A, Shinzawa-Ito, K, Yoshikawa, S, Tsukihara, T. | | Deposit date: | 2016-03-11 | | Release date: | 2017-03-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Bovine heart cytochrome c oxidase in the carbon monoxide-bound mixed-valence state at 1.68 angstrom resolution (50 K)
To Be Published
|
|
5Z84
 
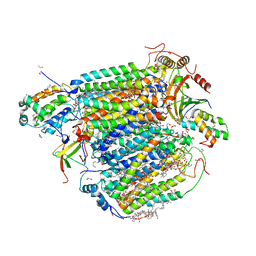 | | The structure of azide-bound cytochrome c oxidase determined using the crystals exposed to 20 mM azide solution for 4 days | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Shimada, A, Hatano, K, Tadehara, H, Tsukihara, T. | | Deposit date: | 2018-01-31 | | Release date: | 2018-08-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | X-ray structural analyses of azide-bound cytochromecoxidases reveal that the H-pathway is critically important for the proton-pumping activity.
J. Biol. Chem., 293, 2018
|
|
5Z85
 
 | | The structure of azide-bound cytochrome c oxidase determined using the another batch crystals exposed to 20 mM azide solution for 2 days | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Shimada, A, Hatano, K, Tadehara, H, Tsukihara, T. | | Deposit date: | 2018-01-31 | | Release date: | 2018-08-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | X-ray structural analyses of azide-bound cytochromecoxidases reveal that the H-pathway is critically important for the proton-pumping activity.
J. Biol. Chem., 293, 2018
|
|
5ZCP
 
 | | azide-bound cytochrome c oxidase structure determined using the crystals exposed to 20 mM azide solution for 2 days | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Shimada, A, Hatano, K, Tadehara, H, Tsukihara, T. | | Deposit date: | 2018-02-19 | | Release date: | 2018-08-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | X-ray structural analyses of azide-bound cytochromecoxidases reveal that the H-pathway is critically important for the proton-pumping activity.
J. Biol. Chem., 293, 2018
|
|
5Z86
 
 | | azide-bound cytochrome c oxidase structure determined using the crystals exposed to 20 mM azide solution for 3 days | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Shimada, A, Hatano, K, Tadehara, H, Tsukihara, T. | | Deposit date: | 2018-01-31 | | Release date: | 2018-08-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | X-ray structural analyses of azide-bound cytochromecoxidases reveal that the H-pathway is critically important for the proton-pumping activity.
J. Biol. Chem., 293, 2018
|
|
5ZCQ
 
 | | Azide-bound cytochrome c oxidase structure determined using the crystals exposed to 10 mM azide solution for 2 days | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Shimada, A, Hatano, K, Tadehara, H, Tsukihara, T. | | Deposit date: | 2018-02-19 | | Release date: | 2018-08-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | X-ray structural analyses of azide-bound cytochromecoxidases reveal that the H-pathway is critically important for the proton-pumping activity.
J. Biol. Chem., 293, 2018
|
|
7EV7
 
 | | Bovine heart cytochrome c oxidase in the carbon monoxide-bound fully reduced state at a 50 K | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Shimada, A, Yoshikawa, S, Tsukihara, T. | | Deposit date: | 2021-05-20 | | Release date: | 2021-07-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of CO-bound fully reduced state of bovine heart cytochrome c oxidase
To Be Published
|
|
2EFK
 
 | | Crystal structure of the EFC domain of Cdc42-interacting protein 4 | | Descriptor: | Cdc42-interacting protein 4 | | Authors: | Shimada, A, Niwa, H, Chen, L, Liu, Z.-J, Wang, B.-C, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-02-23 | | Release date: | 2007-05-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Curved EFC/F-BAR-Domain Dimers Are Joined End to End into a Filament for Membrane Invagination in Endocytosis
Cell(Cambridge,Mass.), 129, 2007
|
|
2EFL
 
 | | Crystal structure of the EFC domain of formin-binding protein 17 | | Descriptor: | Formin-binding protein 1 | | Authors: | Shimada, A, Niwa, H, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-02-23 | | Release date: | 2007-05-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Curved EFC/F-BAR-Domain Dimers Are Joined End to End into a Filament for Membrane Invagination in Endocytosis
Cell(Cambridge,Mass.), 129, 2007
|
|
6JUW
 
 | | BOVINE HEART CYTOCHROME C OXIDASE IN CATALITIC INTERMEDIATES AT 1.80 ANGSTROM RESOLUTION | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Shimada, A, Muramoto, K, Shinzawa-Itoh, K, Yoshikawa, S, Tsukihara, T. | | Deposit date: | 2019-04-15 | | Release date: | 2020-03-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray structures of catalytic intermediates of cytochromecoxidase provide insights into its O2activation and unidirectional proton-pump mechanisms.
J.Biol.Chem., 295, 2020
|
|
