2V9A
 
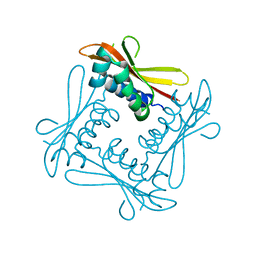 | | Structure of Citrate-free Periplasmic Domain of Sensor Histidine Kinase CitA | | Descriptor: | SENSOR KINASE CITA | | Authors: | Sevvana, M, Vijayan, V, Zweckstetter, M, Reinelt, S, Madden, D.R, Sheldrick, G.M, Bott, M, Griesinger, C, Becker, S. | | Deposit date: | 2007-08-23 | | Release date: | 2008-03-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Ligand-Induced Switch in the Periplasmic Domain of Sensor Histidine Kinase Cita.
J.Mol.Biol., 377, 2008
|
|
2J80
 
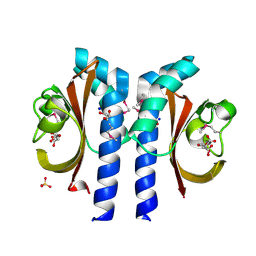 | | Structure of Citrate-bound Periplasmic Domain of Sensor Histidine Kinase CitA | | Descriptor: | CITRATE ANION, GLYCEROL, SENSOR KINASE CITA, ... | | Authors: | Sevvana, M, Vijayan, V, Zweckstetter, M, Reinelt, S, Madden, D.R, Sheldrick, G.M, Bott, M, Griesinger, C, Becker, S. | | Deposit date: | 2006-10-18 | | Release date: | 2007-10-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A Ligand-Induced Switch in the Periplasmic Domain of Sensor Histidine Kinase Cita.
J.Mol.Biol., 377, 2008
|
|
2C0F
 
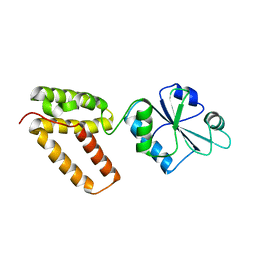 | | Structure of Wind Y53F mutant | | Descriptor: | WINDBEUTEL PROTEIN | | Authors: | Sevvana, M, Ma, Q, Barnewitz, K, Guo, C, Soling, H.-D, Ferrari, D.M, Sheldrick, G.M. | | Deposit date: | 2005-09-02 | | Release date: | 2006-08-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural Elucidation of the Pdi-Related Chaperone Wind with the Help of Mutants.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2C0G
 
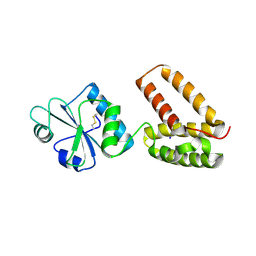 | | Structure of PDI-related Chaperone, Wind mutant-Y53S | | Descriptor: | CHLORIDE ION, SODIUM ION, WINDBEUTEL PROTEIN | | Authors: | Sevvana, M, Ma, Q, Barnewitz, K, Guo, C, Soling, H.-D, Ferrari, D.M, Sheldrick, G.M. | | Deposit date: | 2005-09-02 | | Release date: | 2006-08-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Elucidation of the Pdi-Related Chaperone Wind with the Help of Mutants.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2C1Y
 
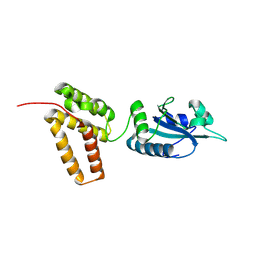 | | Structure of PDI-related Chaperone, Wind mutant-Y55K | | Descriptor: | WINDBEUTEL PROTEIN | | Authors: | Sevvana, M, Ma, Q, Barnewitz, K, Guo, C, Soling, H.-D, Ferrari, D.M, Sheldrick, G.M. | | Deposit date: | 2005-09-22 | | Release date: | 2006-08-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural Elucidation of the Pdi-Related Chaperone Wind with the Help of Mutants.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
5MQQ
 
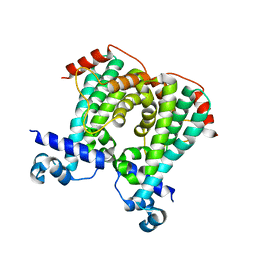 | |
2C0E
 
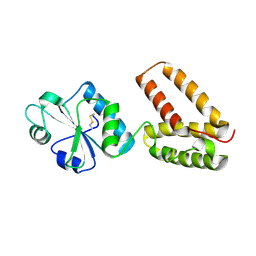 | | Structure of PDI-related Chaperone, Wind with his-tag on C-terminus. | | Descriptor: | WINDBEUTEL PROTEIN | | Authors: | Sevvana, M, Ma, Q, Barnewitz, K, Guo, C, Soling, H.-D, Ferrari, D.M, Sheldrick, G.M. | | Deposit date: | 2005-09-01 | | Release date: | 2006-08-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural Elucidation of the Pdi-Related Chaperone Wind with the Help of Mutants.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
6CO8
 
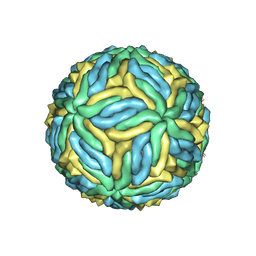 | | Structure of Zika virus at a resolution of 3.1 Angstrom | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, E protein, M protein | | Authors: | Sevvana, M, Long, F, Miller, A.J, Klose, T, Buda, G, Sun, L, Kuhn, R.J, Rossmann, M.R. | | Deposit date: | 2018-03-12 | | Release date: | 2018-07-04 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Refinement and Analysis of the Mature Zika Virus Cryo-EM Structure at 3.1 angstrom Resolution.
Structure, 26, 2018
|
|
6OQZ
 
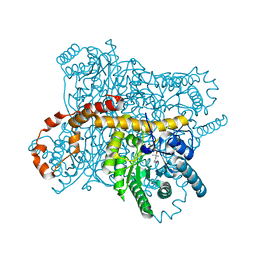 | | Crystal structure of Glucose Isomerase from Non-merohedrally twinned crystals | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Sevvana, M, Ruf, M, Uson, I, Sheldrick, G.M, Herbst-Irmer, R. | | Deposit date: | 2019-04-29 | | Release date: | 2019-12-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Non-merohedral twinning: from minerals to proteins.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6OR0
 
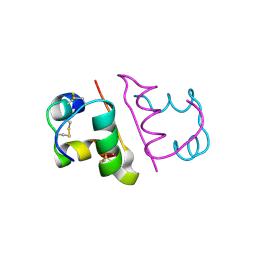 | | Crystal structure of Insulin from Non-merohedrally twinned crystals | | Descriptor: | Insulin chain A, Insulin chain B | | Authors: | Sevvana, M, Ruf, M, Uson, I, Sheldrick, G.M, Herbst-Irmer, R. | | Deposit date: | 2019-04-29 | | Release date: | 2019-12-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Non-merohedral twinning: from minerals to proteins.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
7KCR
 
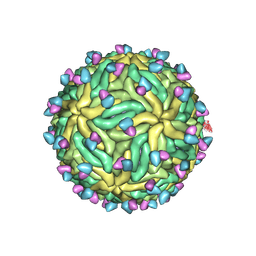 | | Cryo-EM structure of Zika virus in complex with E protein cross-linking human monoclonal antibody ADI30056 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ADI30056 Fab heavy chain variable region, ADI30056 Fab light chain variable region, ... | | Authors: | Sevvana, M, Rogers, T.F, Miller, A.S, Long, F, Klose, T, Beutler, N, Lai, Y.C, Parren, M, Walker, L.M, Buda, G, Burton, D.R, Rossmann, M.G, Kuhn, R.J. | | Deposit date: | 2020-10-07 | | Release date: | 2020-12-16 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural Basis of Zika Virus Specific Neutralization in Subsequent Flavivirus Infections.
Viruses, 12, 2020
|
|
2WEX
 
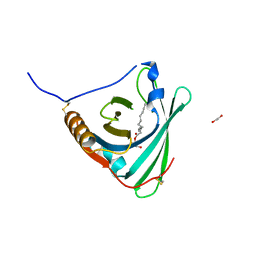 | | Crystal structure of human apoM in complex with glycerol 1- myristic acid | | Descriptor: | (2R)-2,3-dihydroxypropyl tetradecanoate, 1,2-ETHANEDIOL, APOLIPOPROTEIN M | | Authors: | Sevvana, M, Ahnstrom, J, Egerer-Sieber, C, Dahlback, B, Muller, Y.A. | | Deposit date: | 2009-04-02 | | Release date: | 2009-09-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Serendipitous Fatty Acid Binding Reveals the Structural Determinants for Ligand Recognition in Apolipoprotein M.
J.Mol.Biol., 393, 2009
|
|
2WEW
 
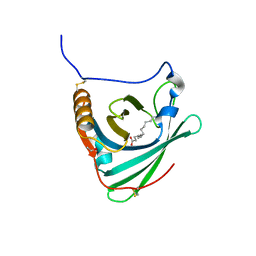 | | Crystal structure of human apoM in complex with myristic acid | | Descriptor: | 1,2-ETHANEDIOL, APOLIPOPROTEIN M, MYRISTIC ACID | | Authors: | Sevvana, M, Ahnstrom, J, Egerer-Sieber, C, Dahlback, B, Muller, Y.A. | | Deposit date: | 2009-04-02 | | Release date: | 2009-09-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Serendipitous Fatty Acid Binding Reveals the Structural Determinants for Ligand Recognition in Apolipoprotein M.
J.Mol.Biol., 393, 2009
|
|
3ZQG
 
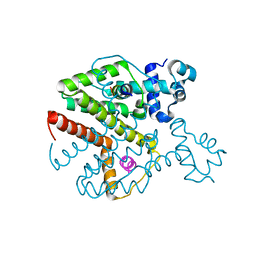 | | Structure of Tetracycline repressor in complex with antiinducer peptide-TAP2 | | Descriptor: | ANTI-INDUCER PEPTIDE TAP2, TETRACYCLINE REPRESSOR PROTEIN CLASS B FROM TRANSPOSON TN10, TETRACYCLINE REPRESSOR PROTEIN CLASS D | | Authors: | Sevvana, M, Goeke, D, Stoeckle, C, Kaspar, D, Grubmueller, S, Goetz, C, Wimmer, C, Berens, C, Klotzsche, M, Muller, Y.A, Hillen, W. | | Deposit date: | 2011-06-09 | | Release date: | 2011-12-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | An Exclusive Alpha/Beta Code Directs Allostery in Tetr-Peptide Complexes.
J.Mol.Biol., 416, 2012
|
|
3ZQI
 
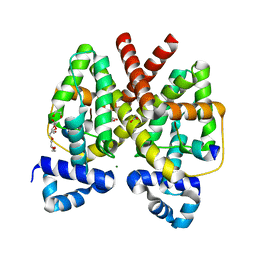 | | Structure of Tetracycline repressor in complex with inducer peptide- TIP2 | | Descriptor: | 1,2-ETHANEDIOL, INDUCER PEPTIDE TIP2, MAGNESIUM ION, ... | | Authors: | Sevvana, M, Goeke, D, Stoeckle, C, Kaspar, D, Grubmueller, S, Goetz, C, Wimmer, C, Berens, C, Klotzsche, M, Muller, Y.A, Hillen, W. | | Deposit date: | 2011-06-09 | | Release date: | 2011-12-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | An Exclusive Alpha/Beta Code Directs Allostery in Tetr-Peptide Complexes.
J.Mol.Biol., 416, 2012
|
|
3ZQH
 
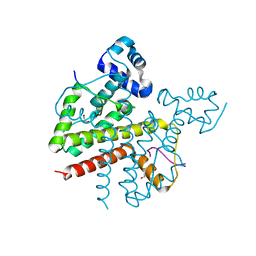 | | Structure of Tetracycline repressor in complex with inducer peptide- TIP3 | | Descriptor: | 1,2-ETHANEDIOL, INDUCER PEPTIDE TIP3, TETRACYCLINE REPRESSOR PROTEIN CLASS B FROM TRANSPOSON TN10, ... | | Authors: | Sevvana, M, Goeke, D, Stoeckle, C, Kaspar, D, Grubmueller, S, Goetz, C, Wimmer, C, Berens, C, Klotzsche, M, Muller, Y.A, Hillen, W. | | Deposit date: | 2011-06-09 | | Release date: | 2011-12-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | An Exclusive Alpha/Beta Code Directs Allostery in Tetr-Peptide Complexes.
J.Mol.Biol., 416, 2012
|
|
3ZQF
 
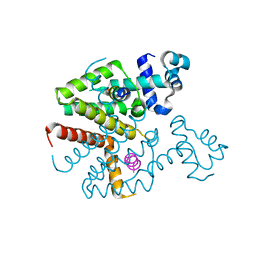 | | Structure of Tetracycline repressor in complex with antiinducer peptide-TAP1 | | Descriptor: | ANTI-INDUCER PEPTIDE TAP1, TETRACYCLINE REPRESSOR PROTEIN CLASS B FROM TRANSPOSON TN10, TETRACYCLINE REPRESSOR PROTEIN CLASS D | | Authors: | Sevvana, M, Goeke, D, Stoeckle, C, Kaspar, D, Grubmueller, S, Goetz, C, Wimmer, C, Berens, C, Klotzsche, M, Muller, Y.A, Hillen, W. | | Deposit date: | 2011-06-09 | | Release date: | 2011-12-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | An Exclusive Alpha/Beta Code Directs Allostery in Tetr-Peptide Complexes.
J.Mol.Biol., 416, 2012
|
|
2XKL
 
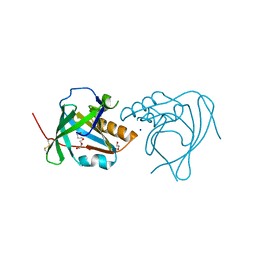 | | Crystal Structure of Mouse Apolipoprotein M | | Descriptor: | 1,2-ETHANEDIOL, APOLIPOPROTEIN M, GLYCEROL, ... | | Authors: | Sevvana, M, Kassler, K, Josefin, A, Weiler, S, Dahlback, B, Sticht, H, Muller, Y.A. | | Deposit date: | 2010-07-09 | | Release date: | 2010-10-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mouse Apom Displays an Unprecedented Seven-Stranded Lipocalin Fold: Folding Decoy or Alternative Native Fold?
J.Mol.Biol., 404, 2010
|
|
6GY3
 
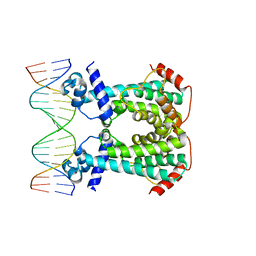 | | Crystal Structure of C. glutamicum AmtR bound to glnA operator DNA | | Descriptor: | AmtR protein, DNA (5'-D(*GP*TP*CP*TP*AP*TP*AP*GP*AP*TP*CP*GP*AP*TP*AP*GP*AP*C)-3'), DNA (5'-D(*GP*TP*CP*TP*AP*TP*CP*GP*AP*TP*CP*TP*AP*TP*AP*GP*AP*C)-3') | | Authors: | Sevvana, M, Muller, Y.A. | | Deposit date: | 2018-06-28 | | Release date: | 2019-07-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | The role of DNA flexibility in transcription regulator AmtR-DNA interaction
To be published
|
|
4WID
 
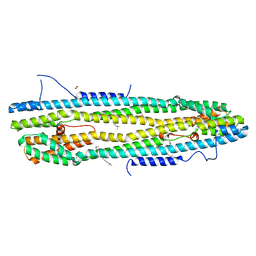 | | Crystal structure of the immediate-early 1 protein (IE1) at 2.31 angstrom (tetragonal form after crystal dehydration) | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, RhUL123 | | Authors: | Klingl, S, Scherer, M, Sevvana, M, Muller, Y.A, Stamminger, T. | | Deposit date: | 2014-09-25 | | Release date: | 2014-10-29 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Crystal Structure of Cytomegalovirus IE1 Protein Reveals Targeting of TRIM Family Member PML via Coiled-Coil Interactions.
Plos Pathog., 10, 2014
|
|
2VKV
 
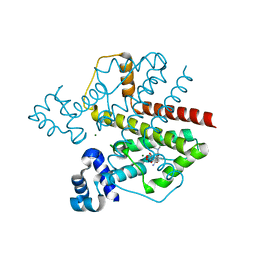 | | TetR (BD) variant L17G with reverse phenotype | | Descriptor: | 5A,6-ANHYDROTETRACYCLINE, MAGNESIUM ION, TETRACYCLINE REPRESSOR PROTEIN CLASS B FROM TRANSPOSON TN10, ... | | Authors: | Resch, M, Striegl, H, Henssler, E.M, Sevvana, M, Egerer-Sieber, C, Schiltz, E, Hillen, W, Muller, Y.A. | | Deposit date: | 2008-01-02 | | Release date: | 2008-07-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | A Protein Functional Leap: How a Single Mutation Reverses the Function of the Transcription Regulator Tetr.
Nucleic Acids Res., 36, 2008
|
|
2QC7
 
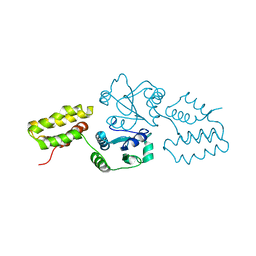 | | Crystal structure of the protein-disulfide isomerase related chaperone ERp29 | | Descriptor: | Endoplasmic reticulum protein ERp29 | | Authors: | Barak, N.N, Sevvana, M, Neumann, P, Malesevic, M, Naumann, K, Fischer, G, Sheldrick, G.M, Stubbs, M.T, Ferrari, D.M. | | Deposit date: | 2007-06-19 | | Release date: | 2008-06-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure and functional analysis of the protein disulfide isomerase-related protein ERp29.
J.Mol.Biol., 385, 2009
|
|
5D5N
 
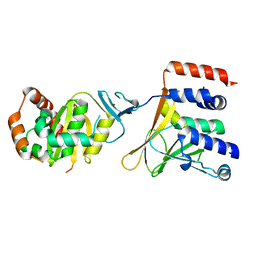 | | Crystal Structure of the Human Cytomegalovirus pUL50-pUL53 Complex | | Descriptor: | Virion egress protein UL31 homolog, Virion egress protein UL34 homolog, ZINC ION | | Authors: | Walzer, S.A, Egerer-Sieber, C, Hohl, K, Sevvana, M, Muller, Y.A. | | Deposit date: | 2015-08-11 | | Release date: | 2015-10-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Crystal Structure of the Human Cytomegalovirus pUL50-pUL53 Core Nuclear Egress Complex Provides Insight into a Unique Assembly Scaffold for Virus-Host Protein Interactions.
J.Biol.Chem., 290, 2015
|
|
8FE3
 
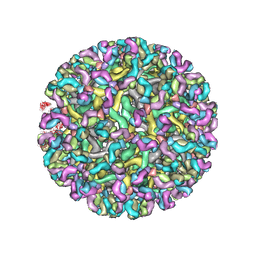 | | Structure of dengue virus (DENV2) in complex with prM12, an anti-PrM monoclonal antibody | | Descriptor: | Envelope protein E, prM protein, prM12 Fab Heavy Chain, ... | | Authors: | Dowd, A.D, Sirohi, D, Speer, S, Mukherjee, S, Govero, J, Aleshnick, M, Larman, B, Sukupolvi-Petty, S, Sevvana, M, Miller, A.S, Klose, T, Zheng, A, Kielian, M, Kuhn, R.J, Diamond, M.S, Pierson, T.C. | | Deposit date: | 2022-12-05 | | Release date: | 2023-02-01 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (10.2 Å) | | Cite: | prM-reactive antibodies reveal a role for partially mature virions in dengue virus pathogenesis.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8FE4
 
 | | Structure of dengue virus (DENV2) in complex with prM13, an anti-PrM monoclonal antibody | | Descriptor: | Envelope protein E, prM protein, prM13 Fab Heavy Chain, ... | | Authors: | Dowd, A.D, Sirohi, D, Speer, S, Mukherjee, S, Govero, J, Aleshnick, M, Larman, B, Sukupolvi-Petty, S, Sevvana, M, Miller, A.S, Klose, T, Zheng, A, Kielian, M, Kuhn, R.J, Diamond, M.S, Pierson, T.C. | | Deposit date: | 2022-12-05 | | Release date: | 2023-02-01 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (9.8 Å) | | Cite: | prM-reactive antibodies reveal a role for partially mature virions in dengue virus pathogenesis.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
