4P5P
 
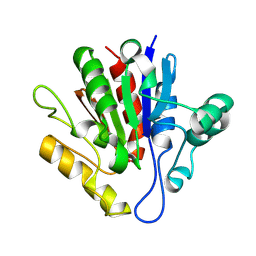 | | X-ray structure of Francisella tularensis Rapid Encystment Protein 24 KDa (REP24), gene product of FTN_0841 | | Descriptor: | ThiJ/PfpI family protein | | Authors: | Segelke, B.W, Feld, G.K, Corzett, M.H, Omattage, N.S, Rasley, A. | | Deposit date: | 2014-03-19 | | Release date: | 2015-04-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | X-ray structure of Francisella tularensis Rapid Encystment Protein 24 KDa (REP24), gene product of FTN_0841
To Be Published
|
|
1A3D
 
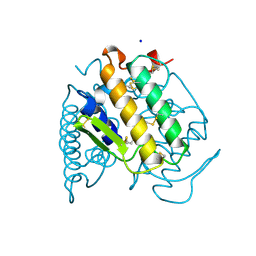 | | PHOSPHOLIPASE A2 (PLA2) FROM NAJA NAJA VENOM | | Descriptor: | PHOSPHOLIPASE A2, SODIUM ION | | Authors: | Segelke, B.W, Nguyen, D, Chee, R, Xuong, H.N, Dennis, E.A. | | Deposit date: | 1998-01-20 | | Release date: | 1998-04-29 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of two novel crystal forms of Naja naja naja phospholipase A2 lacking Ca2+ reveal trimeric packing.
J.Mol.Biol., 279, 1998
|
|
1A3F
 
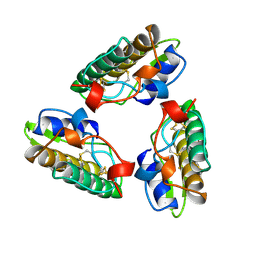 | | PHOSPHOLIPASE A2 (PLA2) FROM NAJA NAJA VENOM | | Descriptor: | PHOSPHOLIPASE A2 | | Authors: | Segelke, B.W, Nguyen, D, Chee, R, Xuong, H.N, Dennis, E.A. | | Deposit date: | 1998-01-21 | | Release date: | 1998-04-29 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structures of two novel crystal forms of Naja naja naja phospholipase A2 lacking Ca2+ reveal trimeric packing.
J.Mol.Biol., 279, 1998
|
|
1OR3
 
 | | APOLIPOPROTEIN E3 (APOE3), TRIGONAL TRUNCATION MUTANT 165 | | Descriptor: | PROTEIN (APOLIPOPROTEIN E) | | Authors: | Rupp, B, Segelke, B.W. | | Deposit date: | 1998-12-01 | | Release date: | 2000-05-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Conformational flexibility in the apolipoprotein E amino-terminal domain structure determined from three new crystal forms: implications for lipid binding.
Protein Sci., 9, 2000
|
|
1OR2
 
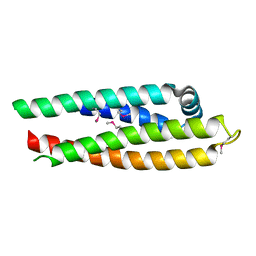 | | APOLIPOPROTEIN E3 (APOE3) TRUNCATION MUTANT 165 | | Descriptor: | APOLIPOPROTEIN E | | Authors: | Rupp, B, Segelke, B.W, Forstner, M. | | Deposit date: | 1999-03-25 | | Release date: | 2000-04-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Conformational flexibility in the apolipoprotein E amino-terminal domain structure determined from three new crystal forms: implications for lipid binding.
Protein Sci., 9, 2000
|
|
3B4T
 
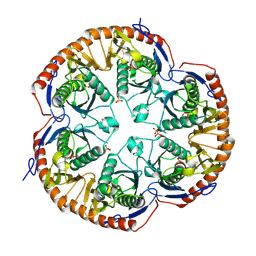 | | Crystal structure of Mycobacterium tuberculosis RNase PH, the Mycobacterium tuberculosis Structural Genomics Consortium target Rv1340 | | Descriptor: | PHOSPHATE ION, Ribonuclease PH | | Authors: | Antczak, A.J, Berger, J.M, Lekin, T, Segelke, B.W, Mycobacterium Tuberculosis Structural Proteomics Project (XMTB), TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2007-10-24 | | Release date: | 2007-11-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | 2.1 A Crystal structure of RNase PH from Mycobacterium tuberculosis.
To be Published
|
|
2NYX
 
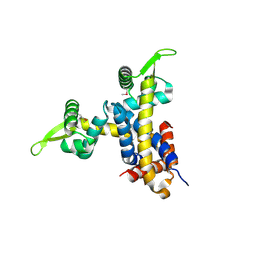 | | Crystal structure of RV1404 from Mycobacterium tuberculosis | | Descriptor: | Probable transcriptional regulatory protein, Rv1404 | | Authors: | Yu, M, Bursey, E.H, Radhakannan, R, Kim, C.-Y, Kaviratne, T, Woodruff, T, Segelke, B.W, Lekin, T, Toppani, D, Terwilliger, T.C, Hung, L.-W, TB Structural Genomics Consortium (TBSGC), Integrated Center for Structure and Function Innovation (ISFI) | | Deposit date: | 2006-11-21 | | Release date: | 2006-12-05 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of RV1404 from Mycobacterium tuberculosis
To be Published
|
|
2A2J
 
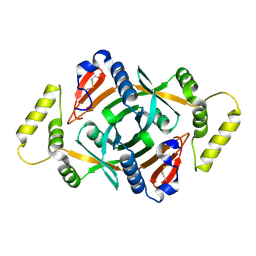 | | Crystal structure of a putative pyridoxine 5'-phosphate oxidase (Rv2607) from Mycobacterium tuberculosis | | Descriptor: | Pyridoxamine 5'-phosphate oxidase | | Authors: | Pedelacq, J.-D, Rho, B.-S, Kim, C.-Y, Waldo, G.S, Lekin, T.P, Segelke, B.W, Rupp, B, Hung, L.-W, Kim, S.-I, Terwilliger, T.C, Mycobacterium Tuberculosis Structural Proteomics Project (XMTB) | | Deposit date: | 2005-06-22 | | Release date: | 2005-08-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a putative pyridoxine 5'-phosphate oxidase (Rv2607) from Mycobacterium tuberculosis.
Proteins, 62, 2005
|
|
4OKO
 
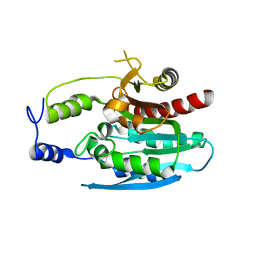 | | Crystal structure of Francisella tularensis REP34 (Rapid Encystment Phenotype Protein 34 KDa) | | Descriptor: | ACETATE ION, Rapid Encystment Phenotype Protein 34 KDa, ZINC ION | | Authors: | Feld, G.K, Segelke, B.W, Corzett, M.H, Hunter, M.S, Frank, M, Rasley, A. | | Deposit date: | 2014-01-22 | | Release date: | 2014-09-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.053 Å) | | Cite: | Structure and Function of REP34 Implicates Carboxypeptidase Activity in Francisella tularensis Host Cell Invasion.
J.Biol.Chem., 289, 2014
|
|
1W30
 
 | | PyrR of Mycobacterium Tuberculosis as a potential drug target | | Descriptor: | PYRR BIFUNCTIONAL PROTEIN | | Authors: | Kantardjieff, K.A, Vasquez, C, Castro, P, Warfel, N.N, Rho, B.-S, Lekin, T, Kim, C.-Y, Segelke, B.W, Terwilliger, T, Rupp, B, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2004-07-11 | | Release date: | 2004-09-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of Pyrr (Rv1379) from Mycobacterium Tuberculosis: A Persistence Gene and Protein Drug Target
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1XVW
 
 | | Crystal Structure of AhpE from Mycobacterium tuberculosis, a 1-Cys peroxiredoxin | | Descriptor: | Hypothetical protein Rv2238c/MT2298 | | Authors: | Li, S, Peterson, N.A, Kim, M.Y, Kim, C.Y, Hung, L.W, Yu, M, Lekin, T, Segelke, B.W, Lott, J.S, Baker, E.N, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2004-10-28 | | Release date: | 2005-02-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of AhpE from Mycobacterium tuberculosis, a 1-Cys Peroxiredoxin
J.Mol.Biol., 346, 2005
|
|
1XXU
 
 | | Crystal Structure of AhpE from Mycrobacterium tuberculosis, a 1-Cys peroxiredoxin | | Descriptor: | Hypothetical protein Rv2238c/MT2298 | | Authors: | Li, S, Peterson, N.A, Kim, M.Y, Kim, C.Y, Hung, L.W, Yu, M, Lekin, T, Segelke, B.W, Lott, J.S, Baker, E.N, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2004-11-08 | | Release date: | 2005-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of AhpE from Mycobacterium tuberculosis, a 1-Cys Peroxiredoxin
J.Mol.Biol., 346, 2005
|
|
1UPI
 
 | | Mycobacterium tuberculosis rmlC epimerase (Rv3465) | | Descriptor: | DTDP-4-DEHYDRORHAMNOSE 3,5-EPIMERASE | | Authors: | Kim, C.-Y, Naranjo, C, Waldo, G.S, Lekin, T, Segelke, B.W, Kantardjieff, K.A, Zemla, A, Terwilliger, T, Rupp, B, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2003-10-05 | | Release date: | 2003-10-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mycobacterium Tuberculosis Rmlc Epimerase (Rv3465): A Promising Drug-Target Structure in the Rhamnose Pathway
Acta Crystallogr.,Sect.D, 60, 2004
|
|
2IB0
 
 | | Crystal structure of a conserved hypothetical protein, rv2844, from Mycobacterium tuberculosis | | Descriptor: | CONSERVED HYPOTHETICAL ALANINE RICH PROTEIN | | Authors: | Yu, M, Bursey, E.H, Radhakannan, T, Kim, C.Y, Kaviratne, T, Woodruff, T, Segelke, B.W, Lekin, T, Toppani, D, Terwilliger, T.C, Hung, L.W, TB Structural Genomics Consortium (TBSGC), Integrated Center for Structure and Function Innovation (ISFI) | | Deposit date: | 2006-09-08 | | Release date: | 2006-09-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a conserved hypothetical protein, rv2844, from Mycobacterium tuberculosis
To be Published
|
|
2FSX
 
 | | Crystal structure of Rv0390 from M. tuberculosis | | Descriptor: | BROMIDE ION, COG0607: Rhodanese-related sulfurtransferase, SULFATE ION | | Authors: | Bursey, E.H, Radhakannan, T, Yu, M, Segelke, B.W, Lekin, T, Toppani, D, Chang, Y.-B, Kaviratne, T, Woodruff, T, Terwilliger, T.C, Hung, L.-W, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2006-01-23 | | Release date: | 2006-02-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Rv0390 from Mycobacterium tuberculosis
To be Published
|
|
2FGG
 
 | | Crystal Structure of Rv2632c | | Descriptor: | Hypothetical protein Rv2632c/MT2708 | | Authors: | Yu, M, Bursey, E.H, Radhakannan, T, Segelke, B.W, Lekin, T, Toppani, D, Kim, C.Y, Kaviratne, T, Woodruff, T, Terwilliger, T.C, Hung, L.W, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2005-12-21 | | Release date: | 2006-02-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Rv2632c
To be Published
|
|
2G2D
 
 | | Crystal structure of a putative pduO-type ATP:cobalamin adenosyltransferase from Mycobacterium tuberculosis | | Descriptor: | ATP:cobalamin adenosyltransferase | | Authors: | Moon, J.H, Kaviratne, A, Yu, M, Bursey, E.H, Hung, L.-W, Lekin, T.P, Segelke, B.W, Terwilliger, T.C, Kim, C.-Y, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2006-02-15 | | Release date: | 2006-03-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a putative pduO-type ATP:cobalamin adenosyltransferase from Mycobacterium tuberculosis.
To be Published
|
|
1DYQ
 
 | | STAPHYLOCOCCAL ENTEROTOXIN A MUTANT VACCINE | | Descriptor: | Enterotoxin type A, SULFATE ION, ZINC ION | | Authors: | Krupka, H.I, Segelke, B.W, Rupp, B. | | Deposit date: | 2000-02-05 | | Release date: | 2001-03-19 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Basis for Abrogated Binding between Staphylococcal Enterotoxin a Superantigen Vaccine and Mhc-II?
Protein Sci., 11, 2002
|
|
1CD1
 
 | |
1HD7
 
 | | A Second Divalent Metal Ion in the Active Site of a New Crystal Form of Human Apurinic/Apyridinimic Endonuclease, Ape1, and its Implications for the Catalytic Mechanism | | Descriptor: | DNA-(APURINIC OR APYRIMIDINIC SITE) LYASE, LEAD (II) ION | | Authors: | Beernink, P.T, Segelke, B.W, Rupp, B. | | Deposit date: | 2000-11-09 | | Release date: | 2001-02-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Two Divalent Metal Ions in the Active Site of a New Crystal Form of Human Apurinic/Apyrimidinic Endonuclease, Ape1: Implications for the Catalytic Mechanism
J.Mol.Biol., 307, 2001
|
|
1E9N
 
 | | A second divalent metal ion in the active site of a new crystal form of human apurinic/apyrimidinic endonuclease, Ape1, and its implications for the catalytic mechanism | | Descriptor: | DNA-(APURINIC OR APYRIMIDINIC SITE) LYASE, LEAD (II) ION | | Authors: | Beernink, P.T, Segelke, B.W, Rupp, B. | | Deposit date: | 2000-10-24 | | Release date: | 2001-02-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Two Divalent Metal Ions in the Active Site of a New Crystal Form of Human Apurinic/Apyrimidinic Endonuclease, Ape1: Implications for the Catalytic Mechanism
J.Mol.Biol., 307, 2001
|
|
1BZ4
 
 | | APOLIPOPROTEIN E3 (APO-E3), TRUNCATION MUTANT 165 | | Descriptor: | PROTEIN (APOLIPOPROTEIN E) | | Authors: | Rupp, B, Segelke, B. | | Deposit date: | 1998-11-05 | | Release date: | 1998-11-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Conformational flexibility in the apolipoprotein E amino-terminal domain structure determined from three new crystal forms: implications for lipid binding.
Protein Sci., 9, 2000
|
|
1E1H
 
 | |
8EKD
 
 | |
1MQ7
 
 | | CRYSTAL STRUCTURE OF DUTPASE FROM MYCOBACTERIUM TUBERCULOSIS (RV2697C) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DEOXYURIDINE 5'-TRIPHOSPHATE NUCLEOTIDOHYDROLASE | | Authors: | Sawaya, M.R, Chan, S, Segelke, B.W, Lekin, T, Heike, K, Cho, U.S, Naranjo, C, Perry, L.J, Yeates, T.O, Eisenberg, D, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2002-09-13 | | Release date: | 2002-10-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of the Mycobacterium tuberculosis dUTPase: insights into the catalytic mechanism.
J.Mol.Biol., 341, 2004
|
|
