1C1Z
 
 | | CRYSTAL STRUCTURE OF HUMAN BETA-2-GLYCOPROTEIN-I (APOLIPOPROTEIN-H) | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, BETA2-GLYCOPROTEIN-I, ... | | Authors: | Schwarzenbacher, R, Zeth, K, Diederichs, K, Gries, A, Kostner, G.M, Laggner, P, Prassl, R. | | Deposit date: | 1999-07-22 | | Release date: | 1999-11-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Crystal structure of human beta2-glycoprotein I: implications for phospholipid binding and the antiphospholipid syndrome.
EMBO J., 18, 1999
|
|
1RCW
 
 | |
3EZQ
 
 | | Crystal Structure of the Fas/FADD Death Domain Complex | | Descriptor: | Protein FADD, SODIUM ION, SULFATE ION, ... | | Authors: | Schwarzenbacher, R, Robinson, H, Stec, B, Riedl, S.J. | | Deposit date: | 2008-10-23 | | Release date: | 2008-12-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | The Fas-FADD death domain complex structure unravels signalling by receptor clustering
Nature, 457, 2009
|
|
2KM6
 
 | | NMR structure of the NLRP7 Pyrin domain | | Descriptor: | NACHT, LRR and PYD domains-containing protein 7 | | Authors: | Pinheiro, A, Proell, M, Schwarzenbacher, R, Peti, W. | | Deposit date: | 2009-07-21 | | Release date: | 2010-06-09 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of the NLRP7 pyrin domain: insight into pyrin-pyrin-mediated effector domain signaling in innate immunity.
J.Biol.Chem., 285, 2010
|
|
3PMD
 
 | | Crystal structure of the sporulation inhibitor pXO1-118 from Bacillus anthracis | | Descriptor: | CHLORIDE ION, Conserved domain protein, UNDECANOIC ACID | | Authors: | Stranzl, G.R, Santelli, E, Bankston, L.A, La Clair, C, Bobkov, A, Schwarzenbacher, R, Godzik, A, Perego, M, Grynberg, M, Liddington, R.C. | | Deposit date: | 2010-11-16 | | Release date: | 2011-01-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural Insights into Inhibition of Bacillus anthracis Sporulation by a Novel Class of Non-heme Globin Sensor Domains.
J.Biol.Chem., 286, 2011
|
|
3PMC
 
 | | Crystal structure of the sporulation inhibitor pXO2-61 from Bacillus anthracis | | Descriptor: | CHLORIDE ION, IODIDE ION, Uncharacterized protein pXO2-61/BXB0075/GBAA_pXO2_0075 | | Authors: | Stranzl, G.R, Santelli, E, Bankston, L.A, La Clair, C, Bobkov, A, Schwarzenbacher, R, Godzik, A, Perego, M, Grynberg, M, Liddington, R.C. | | Deposit date: | 2010-11-16 | | Release date: | 2011-01-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structural Insights into Inhibition of Bacillus anthracis Sporulation by a Novel Class of Non-heme Globin Sensor Domains.
J.Biol.Chem., 286, 2011
|
|
3HLX
 
 | |
3HNH
 
 | | Crystal Structure of PqqC Active Site Mutant Y175S,R179S in complex with a reaction intermediate | | Descriptor: | (2S,7R,9R)-4,5-dihydroxy-2,3,6,7,8,9-hexahydro-1H-pyrrolo[2,3-f]quinoline-2,7,9-tricarboxylic acid, Pyrroloquinoline-quinone synthase | | Authors: | Puehringer, S, Schwarzenbacher, R. | | Deposit date: | 2009-05-31 | | Release date: | 2010-05-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural studies of mutant forms of the PQQ-forming enzyme PqqC in the presence of product and substrate
Proteins, 78, 2010
|
|
3HML
 
 | |
1J7N
 
 | | Anthrax Toxin Lethal factor | | Descriptor: | Lethal Factor precursor, SULFATE ION, ZINC ION | | Authors: | Pannifer, A.D, Wong, T.Y, Schwarzenbacher, R, Renatus, M, Petosa, C, Collier, R.J, Bienkowska, J, Lacy, D.B, Park, S, Leppla, S.H, Hanna, P, Liddington, R.C. | | Deposit date: | 2001-05-17 | | Release date: | 2001-11-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the anthrax lethal factor.
Nature, 414, 2001
|
|
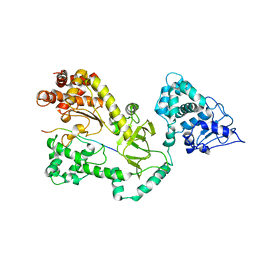
1I3O
 
 | | CRYSTAL STRUCTURE OF THE COMPLEX OF XIAP-BIR2 AND CASPASE 3 | | Descriptor: | BACULOVIRAL IAP REPEAT-CONTAINING PROTEIN 4, CASPASE 3, ZINC ION | | Authors: | Riedl, S.J, Renatus, M, Schwarzenbacher, R, Zhou, Q, Sun, C, Fesik, S.W, Liddington, R.C, Salvesen, G.S. | | Deposit date: | 2001-02-15 | | Release date: | 2001-03-21 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for the inhibition of caspase-3 by XIAP.
Cell(Cambridge,Mass.), 104, 2001
|
|
1JKY
 
 | | Crystal Structure of the Anthrax Lethal Factor (LF): Wild-type LF Complexed with the N-terminal Sequence of MAPKK2 | | Descriptor: | Lethal Factor, mitogen-activated protein kinase kinase 2 | | Authors: | Pannifer, A.D, Wong, T.Y, Schwarzenbacher, R, Renatus, M, Petosa, C, Collier, R.J, Bienkowska, J, Lacy, D.B, Park, S, Leppla, S.H, Hanna, P, Liddington, R.C. | | Deposit date: | 2001-07-13 | | Release date: | 2001-11-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Crystal structure of the anthrax lethal factor.
Nature, 414, 2001
|
|
1YBI
 
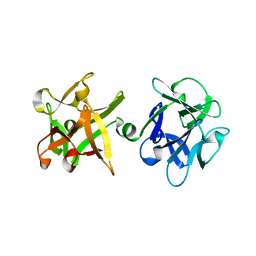 | | Crystal structure of HA33A, a neurotoxin-associated protein from Clostridium botulinum type A | | Descriptor: | non-toxin haemagglutinin HA34 | | Authors: | Arndt, J.W, Gu, J, Jaroszewski, L, Schwarzenbacher, R, Hanson, M, Lebeda, F.J, Stevens, R.C. | | Deposit date: | 2004-12-20 | | Release date: | 2005-02-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Structure of the Neurotoxin-associated Protein HA33/A from Clostridium botulinum Suggests a Reoccurring beta-Trefoil Fold in the Progenitor Toxin Complex.
J.Mol.Biol., 346, 2005
|
|
1Z6T
 
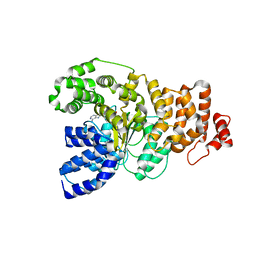 | | Structure of the apoptotic protease-activating factor 1 bound to ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Apoptotic protease activating factor 1 | | Authors: | Riedl, S.J, Li, W, Chao, Y, Schwarzenbacher, R, Shi, Y. | | Deposit date: | 2005-03-23 | | Release date: | 2005-04-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structure of the apoptotic protease-activating factor 1 bound to ADP
Nature, 434, 2005
|
|
1PWP
 
 | | Crystal Structure of the Anthrax Lethal Factor complexed with Small Molecule Inhibitor NSC 12155 | | Descriptor: | Lethal factor, N,N'-BIS(4-AMINO-2-METHYLQUINOLIN-6-YL)UREA, ZINC ION | | Authors: | Wong, T.Y, Schwarzenbacher, R, Liddington, R.C. | | Deposit date: | 2003-07-02 | | Release date: | 2004-01-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Identification of small molecule inhibitors of anthrax lethal factor.
Nat.Struct.Mol.Biol., 11, 2004
|
|
1OTV
 
 | | PqqC, Pyrroloquinolinquinone Synthase C | | Descriptor: | Coenzyme PQQ synthesis protein C | | Authors: | Magnusson, O.T, Toyama, H, Saeki, M, Rojas, A, Reed, J.C, Adachi, O, Klinman, J.P, SChwarzenbacher, R. | | Deposit date: | 2003-03-23 | | Release date: | 2004-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Quinone Biogenesis: Structure and Mechanism of PqqC, the Final Catalyst in the Production of Pyrroloquinoline Quinone.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1OTW
 
 | | Crystal structure of PqqC in complex with PQQ and a putative H2O2 | | Descriptor: | Coenzyme PQQ synthesis protein C, HYDROGEN PEROXIDE, PYRROLOQUINOLINE QUINONE | | Authors: | Magnusson, O.T, Toyama, H, Saeki, M, Rojas, A, Reed, J.C, Liddington, R.C, Klinman, J.P, Schwarzenbacher, R. | | Deposit date: | 2003-03-23 | | Release date: | 2004-05-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Quinone biogenesis: Structure and mechanism of PqqC, the final catalyst in the production of pyrroloquinoline quinone.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1PWQ
 
 | | Crystal structure of Anthrax Lethal Factor complexed with Thioacetyl-Tyr-Pro-Met-Amide, a metal-chelating peptidyl small molecule inhibitor | | Descriptor: | Lethal factor, N-(SULFANYLACETYL)TYROSYLPROLYLMETHIONINAMIDE, ZINC ION | | Authors: | Wong, T.Y, Schwarzenbacher, R, Liddington, R.C. | | Deposit date: | 2003-07-02 | | Release date: | 2004-01-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.52 Å) | | Cite: | The structural basis for substrate and inhibitor selectivity of the anthrax lethal factor.
Nat.Struct.Mol.Biol., 11, 2004
|
|
1PWW
 
 | |
1PWV
 
 | |
1PWU
 
 | | Crystal Structure of Anthrax Lethal Factor complexed with (3-(N-hydroxycarboxamido)-2-isobutylpropanoyl-Trp-methylamide), a known small molecule inhibitor of matrix metalloproteases. | | Descriptor: | 3-(N-HYDROXYCARBOXAMIDO)-2-ISOBUTYLPROPANOYL-TRP-METHYLAMIDE, Lethal factor, ZINC ION | | Authors: | Wong, T.Y, Schwarzenbacher, R, Liddington, R.C. | | Deposit date: | 2003-07-02 | | Release date: | 2004-02-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The structural basis for substrate and inhibitor selectivity of the anthrax lethal factor.
Nat.Struct.Mol.Biol., 11, 2004
|
|
2B8N
 
 | |
1J5S
 
 | |
1O1Y
 
 | |
1O4T
 
 | |
