2KO2
 
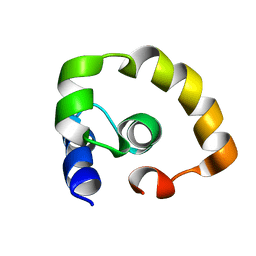 | | NOGO66 | | Descriptor: | Reticulon-4 | | Authors: | Cocco, M.J, Schulz, J, Vasudevan, S.V. | | Deposit date: | 2009-09-08 | | Release date: | 2010-04-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Protein folding at the membrane interface, the structure of Nogo-66 requires interactions with a phosphocholine surface.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
5AHV
 
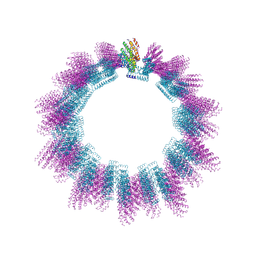 | | Cryo-EM structure of helical ANTH and ENTH tubules on PI(4,5)P2-containing membranes | | Descriptor: | ANTH DOMAIN OF ENDOCYTIC ADAPTOR SLA2, ENTH DOMAIN OF EPSIN ENT1 | | Authors: | Skruzny, M, Desfosses, A, Prinz, S, Dodonova, S.O, Gieras, A, Uetrecht, C, Jakobi, A.J, Abella, M, Hagen, W.J.H, Schulz, J, Meijers, R, Rybin, V, Briggs, J.A.G, Sachse, C, Kaksonen, M. | | Deposit date: | 2015-02-10 | | Release date: | 2015-05-06 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (13.6 Å) | | Cite: | An Organized Co-Assembly of Clathrin Adaptors is Essential for Endocytosis.
Dev.Cell, 33, 2015
|
|
4AC5
 
 | | Lipidic sponge phase crystal structure of the Bl. viridis reaction centre solved using serial femtosecond crystallography | | Descriptor: | 15-cis-1,2-dihydroneurosporene, BACTERIOCHLOROPHYLL B, BACTERIOPHEOPHYTIN B, ... | | Authors: | Johansson, L.C, Arnlund, D, White, T.A, Katona, G, DePonte, D.P, Weierstall, U, Doak, R.B, Shoeman, R.L, Lomb, L, Malmerberg, E, Davidsson, J, Nass, K, Liang, M, Andreasson, J, Aquila, A, Bajt, S, Barthelmess, M, Barty, A, Bogan, M.J, Bostedt, C, Bozek, J.D, Caleman, C, Coffee, R, Coppola, N, Ekeberg, T, Epp, S.W, Erk, B, Fleckenstein, H, Foucar, L, Graafsma, H, Gumprecht, L, Hajdu, J, Hampton, C.Y, Hartmann, R, Hartmann, A, Hauser, G, Hirsemann, H, Holl, P, Hunter, M.S, Kassemeyer, S, Kimmel, N, Kirian, R.A, Maia, F.R.N.C, Marchesini, S, Martin, A.V, Reich, C, Rolles, D, Rudek, B, Rudenko, A, Schlichting, I, Schulz, J, Seibert, M.M, Sierra, R, Soltau, H, Starodub, D, Stellato, F, Stern, S, Struder, L, Timneanu, N, Ullrich, J, Wahlgren, W.Y, Wang, X, Weidenspointner, G, Wunderer, C, Fromme, P, Chapman, H.N, Spence, J.C.H, Neutze, R. | | Deposit date: | 2011-12-14 | | Release date: | 2012-02-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (8.2 Å) | | Cite: | Lipidic Phase Membrane Protein Serial Femtosecond Crystallography.
Nat.Methods, 9, 2012
|
|
3PCQ
 
 | | Femtosecond X-ray protein Nanocrystallography | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Chapman, H.N, Fromme, P, Barty, A, White, T.A, Kirian, R.A, Aquila, A, Hunter, M.S, Schulz, J, Deponte, D.P, Weierstall, U, Doak, R.B, Maia, F.R.N.C, Martin, A.V, Schlichting, I, Lomb, L, Coppola, N, Shoeman, R.L, Epp, S.W, Hartmann, R, Rolles, D, Rudenko, A, Foucar, L, Kimmel, N, Weidenspointner, G, Holl, P, Liang, M, Barthelmess, M, Caleman, C, Boutet, S, Bogan, M.J, Krzywinski, J, Bostedt, C, Bajt, S, Gumprecht, L, Rudek, B, Erk, B, Schmidt, C, Homke, A, Reich, C, Pietschner, D, Struder, L, Hauser, G, Gorke, H, Ullrich, J, Herrmann, S, Schaller, G, Schopper, F, Soltau, H, Kuhnel, K.-U, Messerschmidt, M, Bozek, J.D, Hau-Riege, S.P, Frank, M, Hampton, C.Y, Sierra, R, Starodub, D, Williams, G.J, Hajdu, J, Timneanu, N, Seibert, M.M, Andreasson, J, Rocker, A, Jonsson, O, Svenda, M, Stern, S, Nass, K, Andritschke, R, Schroter, C.-D, Krasniqi, F, Bott, M, Schmidt, K.E, Wang, X, Grotjohann, I, Holton, J.M, Barends, T.R.M, Neutze, R, Marchesini, S, Fromme, R, Schorb, S, Rupp, D, Adolph, M, Gorkhover, T, Andersson, I, Hirsemann, H, Potdevin, G, Graafsma, H, Nilsson, B, Spence, J.C.H. | | Deposit date: | 2010-10-21 | | Release date: | 2011-02-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (8.984 Å) | | Cite: | Femtosecond X-ray protein nanocrystallography.
Nature, 470, 2011
|
|
8QOY
 
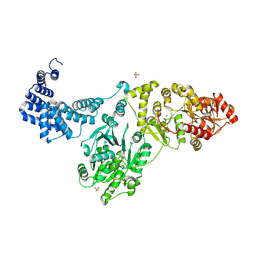 | | Capsular polysaccharide synthesis multienzyme of Actinobacillus Pleuropneumoniae serotype 3 | | Descriptor: | SULFATE ION, TagF-like capsule polymerase Cps3D, ZINC ION | | Authors: | Di Domenico, V, Litschko, C, Schulze, J, Bethe, A, Cifuente, J.O, Marina, A, Budde, I, Mast, T.A, Sulewska, M, Berger, M, Buettner, F, Gerardy-Schahn, R, Fiebig, T, Guerin, M.E. | | Deposit date: | 2023-09-29 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Transition transferases prime bacterial capsule polymerization.
Nat.Chem.Biol., 2024
|
|
8QFG
 
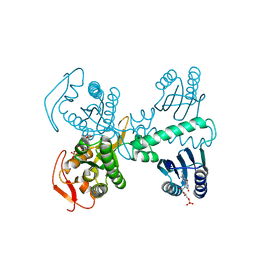 | | Cryogenic crystal structure of the Photoactivated Adenylate Cyclase OaPAC after 5 seconds of blue light illumination | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, FLAVIN MONONUCLEOTIDE, Family 3 adenylate cyclase, ... | | Authors: | Chretien, A, Nagel, M.F, Botha, S, de Wijn, R, Brings, L, Doerner, K, Han, H, C.P.Koliyadu, J, Letrun, R, Round, A, Sato, T, Schmidt, C, Secareanu, R, von Stetten, D, Vakili, M, Wrona, A, Bean, R, Mancuso, A, Schulz, J, R.Pearson, A, Kottke, T, Lorenzen, K, Schubert, R. | | Deposit date: | 2023-09-04 | | Release date: | 2023-11-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Light-induced Trp in /Met out Switching During BLUF Domain Activation in ATP-bound Photoactivatable Adenylate Cyclase OaPAC.
J.Mol.Biol., 436, 2024
|
|
8QFF
 
 | | Cryogenic crystal structure of the Photoactivated Adenylate Cyclase OaPAC with ATP bound | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, FLAVIN MONONUCLEOTIDE, Family 3 adenylate cyclase, ... | | Authors: | Chretien, A, Nagel, M.F, Botha, S, de Wijn, R, Brings, L, Doerner, K, Han, H, C.P.Koliyadu, J, Letrun, R, Round, A, Sato, T, Schmidt, C, Secareanu, R, von Stetten, D, Vakili, M, Wrona, A, Bean, R, Mancuso, A, Schulz, J, R.Pearson, A, Kottke, T, Lorenzen, K, Schubert, R. | | Deposit date: | 2023-09-11 | | Release date: | 2023-11-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Light-induced Trp in /Met out Switching During BLUF Domain Activation in ATP-bound Photoactivatable Adenylate Cyclase OaPAC.
J.Mol.Biol., 436, 2024
|
|
8QFE
 
 | | Cryogenic crystal structure of the Photoactivated Adenylate Cyclase OaPAC | | Descriptor: | FLAVIN MONONUCLEOTIDE, Family 3 adenylate cyclase, MAGNESIUM ION | | Authors: | Chretien, A, Nagel, M.F, Botha, S, de Wijn, R, Brings, L, Doerner, K, Han, H, Koliyadu, J, Letrun, R, Round, A, Sato, T, Schmidt, C, Secareanu, R, von Stetten, D, Vakili, M, Wrona, A, Bean, R, Mancuso, A, Schulz, J, Pearson, A, Kottke, T, Lorenzen, K, Schubert, R. | | Deposit date: | 2023-09-04 | | Release date: | 2023-11-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Light-induced Trp in /Met out Switching During BLUF Domain Activation in ATP-bound Photoactivatable Adenylate Cyclase OaPAC.
J.Mol.Biol., 436, 2024
|
|
8QFH
 
 | | Room temperature crystal structure of the Photoactivated Adenylate Cyclase OaPAC with ATP bound | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, FLAVIN MONONUCLEOTIDE, Family 3 adenylate cyclase, ... | | Authors: | Chretien, A, Nagel, M.F, Botha, S, de Wijn, R, Brings, L, Doerner, K, Han, H, C.P.Koliyadu, J, Letrun, R, Round, A, Sato, T, Schmidt, C, Secareanu, R, von Stetten, D, Vakili, M, Wrona, A, Bean, R, Mancuso, A, Schulz, J, R.Pearson, A, Kottke, T, Lorenzen, K, Schubert, R. | | Deposit date: | 2023-09-04 | | Release date: | 2023-11-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Light-induced Trp in /Met out Switching During BLUF Domain Activation in ATP-bound Photoactivatable Adenylate Cyclase OaPAC.
J.Mol.Biol., 436, 2024
|
|
8QFJ
 
 | | Room temperature crystal structure of the Photoactivated Adenylate Cyclase OaPAC after blue light excitation at 2.3 us delay | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, FLAVIN MONONUCLEOTIDE, Family 3 adenylate cyclase, ... | | Authors: | Chretien, A, Nagel, M.F, Botha, S, de Wijn, R, Brings, L, Doerner, K, Han, H, C.P.Koliyadu, J, Letrun, R, Round, A, Sato, T, Schmidt, C, Secareanu, R, von Stetten, D, Vakili, M, Wrona, A, Bean, R, Mancuso, A, Schulz, J, R.Pearson, A, Kottke, T, Lorenzen, K, Schubert, R. | | Deposit date: | 2023-09-04 | | Release date: | 2023-11-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Light-induced Trp in /Met out Switching During BLUF Domain Activation in ATP-bound Photoactivatable Adenylate Cyclase OaPAC.
J.Mol.Biol., 436, 2024
|
|
8QFI
 
 | | Room temperature crystal structure of the Photoactivated Adenylate Cyclase OaPAC after blue light excitation at 1.8 us delay | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, FLAVIN MONONUCLEOTIDE, Family 3 adenylate cyclase, ... | | Authors: | Chretien, A, Nagel, M.F, Botha, S, de Wijn, R, Brings, L, Doerner, K, Han, H, C.P.Koliyadu, J, Letrun, R, Sato, T, Round, A, Schmidt, C, Secareanu, R, von Stetten, D, Vakili, M, Wrona, A, Bean, R, Mancuso, A, Schulz, J, R.Pearson, A, Kottke, T, Lorenzen, K, Schubert, R. | | Deposit date: | 2023-09-04 | | Release date: | 2023-11-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Light-induced Trp in /Met out Switching During BLUF Domain Activation in ATP-bound Photoactivatable Adenylate Cyclase OaPAC.
J.Mol.Biol., 436, 2024
|
|
4BGH
 
 | | Crystal Structure of CDK2 in complex with pan-CDK Inhibitor | | Descriptor: | 4-((5-BROMO-4-(PROP-2-YN-1-YLAMINO)PYRIMIDIN-2-YL)AMINO)BENZENESULFONAMIDE, CYCLIN-DEPENDENT KINASE 2 | | Authors: | Luecking, U, Jautelat, R, Krueger, M, Brumby, T, Lienau, P, Schaefer, M, Briem, H, Schulze, J, Hillisch, A, Reichel, A, Siemeister, G. | | Deposit date: | 2013-03-26 | | Release date: | 2013-09-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Lab Oddity Prevails: Discovery of Pan-Cdk Inhibitor (R)- S-Cyclopropyl-S-(4-{[4-{[(1R,2R)-2-Hydroxy-1-Methylpropyl]Oxy}-5-(Trifluoromethyl)Pyrimidin-2-Yl]Amino}Phenyl)Sulfoximide (Bay 1000394) for the Treatment of Cancer.
Chemmedchem, 8, 2013
|
|
8A0C
 
 | | Capsular polysaccharide synthesis multienzyme in complex with CMP | | Descriptor: | Bcs3, CYTIDINE-5'-MONOPHOSPHATE, GLYCEROL, ... | | Authors: | Cifuente, J.O, Schulze, J, Bethe, A, Di Domenico, V, Litschko, C, Budde, I, Eidenberger, L, Thiesler, H, Ramon-Roth, I, Berger, M, Claus, H, DAngelo, C, Marina, A, Gerardy-Schahn, R, Schubert, M, Guerin, M.E, Fiebig, T. | | Deposit date: | 2022-05-27 | | Release date: | 2023-04-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A multi-enzyme machine polymerizes the Haemophilus influenzae type b capsule.
Nat.Chem.Biol., 19, 2023
|
|
8A0M
 
 | | Capsular polysaccharide synthesis multienzyme in complex with capsular polymer fragment | | Descriptor: | Bcs3, MAGNESIUM ION, beta-D-ribosyl-(1->1)-D-ribitol-5-phosphate | | Authors: | Cifuente, J.O, Schulze, J, Bethe, A, Di Domenico, V, Litschko, C, Budde, I, Eidenberger, L, Thiesler, H, Ramon-Roth, I, Berger, M, Claus, H, DAngelo, C, Marina, A, Gerardy-Schahn, R, Schubert, M, Guerin, M.E, Fiebig, T. | | Deposit date: | 2022-05-29 | | Release date: | 2023-04-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | A multi-enzyme machine polymerizes the Haemophilus influenzae type b capsule.
Nat.Chem.Biol., 19, 2023
|
|
6WEB
 
 | | Multi-Hit SFX using MHz XFEL sources | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Holmes, S, Darmanin, C, Abbey, B. | | Deposit date: | 2020-04-01 | | Release date: | 2021-10-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Megahertz pulse trains enable multi-hit serial femtosecond crystallography experiments at X-ray free electron lasers.
Nat Commun, 13, 2022
|
|
6WEC
 
 | | Multi-Hit SFX using MHz XFEL sources | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Holmes, S, Darmanin, C, Abbey, B. | | Deposit date: | 2020-04-01 | | Release date: | 2021-10-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Megahertz pulse trains enable multi-hit serial femtosecond crystallography experiments at X-ray free electron lasers.
Nat Commun, 13, 2022
|
|
3MOR
 
 | | Crystal structure of Cathepsin B from Trypanosoma Brucei | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Cathepsin B-like cysteine protease, ... | | Authors: | Cupelli, K, Stehle, T. | | Deposit date: | 2010-04-23 | | Release date: | 2011-11-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | In vivo protein crystallization opens new routes in structural biology.
Nat.Methods, 9, 2012
|
|
7TUM
 
 | | Multi-Hit SFX using MHz XFEL sources- first hit | | Descriptor: | 1,2-ETHANEDIOL, Lysozyme C, SODIUM ION | | Authors: | Darmanin, C, Holmes, S, Abbey, B. | | Deposit date: | 2022-02-03 | | Release date: | 2022-07-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.202 Å) | | Cite: | Megahertz pulse trains enable multi-hit serial femtosecond crystallography experiments at X-ray free electron lasers.
Nat Commun, 13, 2022
|
|
6U57
 
 | |
8RUS
 
 | | Hen egg-white lysozyme (HEWL) structure from EuXFEL FXE, multi-hit Droplet-on-Demand (DoD) injection, 9.3 keV photon energy, space group P432121 | | Descriptor: | ACETATE ION, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Perrett, S, van Thor, J.J. | | Deposit date: | 2024-01-31 | | Release date: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Kilohertz droplet-on-demand serial femtosecond crystallography at the European XFEL station FXE.
Struct Dyn., 11, 2024
|
|
7PXZ
 
 | | Reduced form of SARS-CoV-2 Main Protease determined by XFEL radiation | | Descriptor: | 3C-like proteinase nsp5, CHLORIDE ION | | Authors: | Schubert, R, Reinke, P, Galchenkova, M, Oberthuer, D, Murillo, G.E.P, Kim, C, Bean, R, Turk, D, Hinrichs, W, Middendorf, P, Round, A, Schmidt, C, Mills, G, Kirkwood, H, Han, H, Koliyadu, J, Bielecki, J, Gelisio, L, Sikorski, M, Kloos, M, Vakilii, M, Yefanov, O.N, Vagovic, P, de-Wijn, R, Letrun, R, Guenther, S, White, T.A, Sato, T, Srinivasan, V, Kim, Y, Chretien, A, Han, S, Brognaro, H, Maracke, J, Knoska, J, Seychell, B.C, Brings, L, Norton-Baker, B, Geng, T, Dore, A.S, Uetrecht, C, Redecke, L, Beck, T, Lorenzen, K, Betzel, C, Mancuso, A.P, Bajt, S, Chapman, H.N, Meents, A, Lane, T.J. | | Deposit date: | 2021-10-08 | | Release date: | 2023-01-18 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | SARS-CoV-2 M pro responds to oxidation by forming disulfide and NOS/SONOS bonds.
Nat Commun, 15, 2024
|
|
7PZQ
 
 | | Oxidized form of SARS-CoV-2 Main Protease determined by XFEL radiation | | Descriptor: | 3C-like proteinase nsp5, DIMETHYL SULFOXIDE | | Authors: | Schubert, R, Reinke, P, Galchenkova, M, Oberthuer, D, Murillo, G.E.P, Kim, C, Bean, R, Turk, D, Hinrichs, W, Middendorf, P, Round, A, Schmidt, C, Mills, G, Kirkwood, H, Han, H, Koliyadu, J, Bielecki, J, Gelisio, L, Sikorski, M, Kloos, M, Vakilii, M, Yefanov, O.N, Vagovic, P, de-Wijn, R, Letrun, R, Guenther, S, White, T.A, Sato, T, Srinivasan, V, Kim, Y, Chretien, A, Han, S, Brognaro, H, Maracke, J, Knoska, J, Seychell, B.C, Brings, L, Norton-Baker, B, Geng, T, Dore, A.S, Uetrecht, C, Redecke, L, Beck, T, Lorenzen, K, Betzel, C, Mancuso, A.P, Bajt, S, Chapman, H.N, Meents, A, Lane, T.J. | | Deposit date: | 2021-10-13 | | Release date: | 2023-01-25 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | SARS-CoV-2 M pro responds to oxidation by forming disulfide and NOS/SONOS bonds.
Nat Commun, 15, 2024
|
|
6GTH
 
 | | Serial Femtosecond Crystallography at Megahertz pulse rates | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, Beta-lactamase | | Authors: | Wiedorn, M, Oberthuer, D, Werner, N, Schubert, R, White, T.A, Mancuso, A, Perbandt, M, Betzel, C, Barty, A, Chapman, H. | | Deposit date: | 2018-06-18 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Megahertz serial crystallography.
Nat Commun, 9, 2018
|
|
6FTR
 
 | | Serial Femtosecond Crystallography at Megahertz pulse rates | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Wiedorn, M.O, Oberthuer, D, Barty, A, Chapman, H.N. | | Deposit date: | 2018-02-23 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.76000106 Å) | | Cite: | Megahertz serial crystallography.
Nat Commun, 9, 2018
|
|
8TU3
 
 | | Bruton's tyrosine kinase in complex with covalent inhibitor compound 10 | | Descriptor: | 1-[(4R)-4-{[(6P,8S)-6-(1-methyl-1H-pyrazol-4-yl)pyrazolo[1,5-a]pyrazin-4-yl]oxy}azepan-1-yl]propan-1-one, Tyrosine-protein kinase BTK | | Authors: | Metrick, C.M. | | Deposit date: | 2023-08-15 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery and Preclinical Characterization of BIIB129, a Covalent, Selective, and Brain-Penetrant BTK Inhibitor for the Treatment of Multiple Sclerosis.
J.Med.Chem., 67, 2024
|
|
