7ZJP
 
 | | Optimization of TEAD P-Site Binding Fragment Hit into In Vivo Active Lead MSC-4106 | | Descriptor: | 2-methyl-4-[4-(trifluoromethyl)phenyl]pyrazolo[3,4-b]indole-7-carboxylic acid, SULFATE ION, Transcriptional enhancer factor TEF-1 | | Authors: | Freire, F, Heinrich, T, Petersson, C, Schneider, R, Garg, S, Schwarz, D, Gunera, J, Seshire, A, Koetzner, L, Schlesiger, S, Musil, D, Schilke, H, Doerfel, B, Diehl, P, Boepple, P, Lemos, A.R, Sousa, P.M.F, Freire, F, Bandeiras, T.M, Carswell, E, Pearson, N, Sirohi, S, Hooker, M, Trivier, E, Broome, R, Balsiger, A, Crowden, A, Dillon, C, Wienke, D. | | Deposit date: | 2022-04-11 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Optimization of TEAD P-Site Binding Fragment Hit into In Vivo Active Lead MSC-4106 .
J.Med.Chem., 65, 2022
|
|
2UVS
 
 | | High Resolution Solid-state NMR structure of Kaliotoxin | | Descriptor: | POTASSIUM CHANNEL TOXIN ALPHA-KTX 3.1 | | Authors: | Korukottu, J, Lange, A, Vijayan, V, Schneider, R, Pongs, O, Becker, S, Baldus, M, Zweckstetter, M. | | Deposit date: | 2007-03-14 | | Release date: | 2008-05-27 | | Last modified: | 2024-11-06 | | Method: | SOLID-STATE NMR | | Cite: | Conformational Plasticity in Ion Channel Recognition of a Peptide Toxin
To be Published
|
|
3ZFN
 
 | | Crystal structure of product-like, processed N-terminal protease Npro | | Descriptor: | CHLORIDE ION, MONOTHIOGLYCEROL, N-TERMINAL PROTEASE NPRO | | Authors: | Zogg, T, Sponring, M, Schindler, S, Koll, M, Schneider, R, Brandstetter, H, Auer, B. | | Deposit date: | 2012-12-12 | | Release date: | 2013-05-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structures of the Viral Protease Npro Imply Distinct Roles for the Catalytic Water in Catalysis
Structure, 21, 2013
|
|
3ZFQ
 
 | | Crystal structure of product-like, processed N-terminal protease Npro with mercury | | Descriptor: | MERCURY (II) ION, MONOTHIOGLYCEROL, N-TERMINAL PROTEASE NPRO | | Authors: | Zogg, T, Sponring, M, Schindler, S, Koll, M, Schneider, R, Brandstetter, H, Auer, B. | | Deposit date: | 2012-12-12 | | Release date: | 2013-05-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal Structures of the Viral Protease Npro Imply Distinct Roles for the Catalytic Water in Catalysis
Structure, 21, 2013
|
|
3ZFT
 
 | | Crystal structure of product-like, processed N-terminal protease Npro at pH 3 | | Descriptor: | CHLORIDE ION, MONOTHIOGLYCEROL, N-TERMINAL PROTEASE NPRO | | Authors: | Zogg, T, Sponring, M, Schindler, S, Koll, M, Schneider, R, Brandstetter, H, Auer, B. | | Deposit date: | 2012-12-12 | | Release date: | 2013-05-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of the Viral Protease Npro Imply Distinct Roles for the Catalytic Water in Catalysis
Structure, 21, 2013
|
|
3ZFO
 
 | | Crystal structure of substrate-like, unprocessed N-terminal protease Npro mutant S169P | | Descriptor: | CHLORIDE ION, HYDROXIDE ION, MONOTHIOGLYCEROL, ... | | Authors: | Zogg, T, Sponring, M, Schindler, S, Koll, M, Schneider, R, Brandstetter, H, Auer, B. | | Deposit date: | 2012-12-12 | | Release date: | 2013-05-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal Structures of the Viral Protease Npro Imply Distinct Roles for the Catalytic Water in Catalysis
Structure, 21, 2013
|
|
3ZFP
 
 | | Crystal structure of product-like, processed N-terminal protease Npro with internal His-Tag | | Descriptor: | CHLORIDE ION, MONOTHIOGLYCEROL, N-TERMINAL PROTEASE NPRO | | Authors: | Zogg, T, Sponring, M, Schindler, S, Koll, M, Schneider, R, Brandstetter, H, Auer, B. | | Deposit date: | 2012-12-12 | | Release date: | 2013-05-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal Structures of the Viral Protease Npro Imply Distinct Roles for the Catalytic Water in Catalysis
Structure, 21, 2013
|
|
3ZFU
 
 | | Crystal structure of substrate-like, unprocessed N-terminal protease Npro mutant S169P with sulphate | | Descriptor: | MONOTHIOGLYCEROL, N-TERMINAL PROTEASE NPRO, SULFATE ION | | Authors: | Zogg, T, Sponring, M, Schindler, S, Koll, M, Schneider, R, Brandstetter, H, Auer, B. | | Deposit date: | 2012-12-12 | | Release date: | 2013-05-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal Structures of the Viral Protease Npro Imply Distinct Roles for the Catalytic Water in Catalysis
Structure, 21, 2013
|
|
3ZFR
 
 | | Crystal structure of product-like, processed N-terminal protease Npro with iridium | | Descriptor: | HYDROXIDE ION, IRIDIUM (III) ION, MONOTHIOGLYCEROL, ... | | Authors: | Zogg, T, Sponring, M, Schindler, S, Koll, M, Schneider, R, Brandstetter, H, Auer, B. | | Deposit date: | 2012-12-12 | | Release date: | 2013-05-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of the Viral Protease Npro Imply Distinct Roles for the Catalytic Water in Catalysis
Structure, 21, 2013
|
|
5BNJ
 
 | | CDK8/CYCC IN COMPLEX WITH 8-{3-Chloro-5-[4-(1-methyl-1H-pyrazol-4-yl)-phenyl]-pyridin- 4-yl}-2,8-diaza-spiro[4.5]decan-1-one | | Descriptor: | 8-{3-chloro-5-[4-(1-methyl-1H-pyrazol-4-yl)phenyl]pyridin-4-yl}-2,8-diazaspiro[4.5]decan-1-one, Cyclin-C, Cyclin-dependent kinase 8, ... | | Authors: | Musil, D, Blagg, J, Wienke, D. | | Deposit date: | 2015-05-26 | | Release date: | 2015-10-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | A selective chemical probe for exploring the role of CDK8 and CDK19 in human disease.
Nat.Chem.Biol., 11, 2015
|
|
7PON
 
 | |
7PNO
 
 | | C terminal domain of Nipah Virus Phosphoprotein fused to the Ntail alpha more of the Nucleoprotein. | | Descriptor: | Phosphoprotein, alpha MoRE of Nipah virus Nucleoprotein tail | | Authors: | Bourhis, J.M, Yabukaski, F, Tarbouriech, N, Jamin, M. | | Deposit date: | 2021-09-07 | | Release date: | 2022-04-20 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structural Dynamics of the C-terminal X Domain of Nipah and Hendra Viruses Controls the Attachment to the C-terminal Tail of the Nucleocapsid Protein.
J.Mol.Biol., 434, 2022
|
|
9GAX
 
 | | TEAD1 in complex with a reversible inhibitor N-[(1S)-2-hydroxy-1-(1-methyl-1H-pyrazol-3-yl)ethyl]-2-methyl-8-[4-(trifluoromethyl)phenyl]-2H,8H-pyrazolo[3,4-b]indole-5-carboxamide | | Descriptor: | 2-methyl-~{N}-[(1~{S})-1-(1-methylpyrazol-3-yl)-2-oxidanyl-ethyl]-4-[4-(trifluoromethyl)phenyl]pyrazolo[3,4-b]indole-7-carboxamide, GLYCEROL, SULFATE ION, ... | | Authors: | Musil, D, Freire, F. | | Deposit date: | 2024-07-29 | | Release date: | 2025-01-22 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | MoA Studies of the TEAD P-Site Binding Ligand MSC-4106 and Its Optimization to TEAD1-Selective Amide M3686.
J.Med.Chem., 68, 2025
|
|
7OCV
 
 | | Human TNKS1 in complex with 3-[4-(1-Hydroxy-1-methyl-ethyl)-phenyl]-6-methyl-2H-pyrrolo[1,2-a]pyrazin-1-one | | Descriptor: | 6-methyl-3-[4-(2-oxidanylpropan-2-yl)phenyl]-4~{H}-pyrrolo[1,2-a]pyrazin-1-one, ACETATE ION, Poly [ADP-ribose] polymerase, ... | | Authors: | Musil, D, Lehmann, M, Buchstaller, H.-P. | | Deposit date: | 2021-04-28 | | Release date: | 2021-07-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.432 Å) | | Cite: | Optimization of a Screening Hit toward M2912, an Oral Tankyrase Inhibitor with Antitumor Activity in Colorectal Cancer Models.
J.Med.Chem., 64, 2021
|
|
5FGK
 
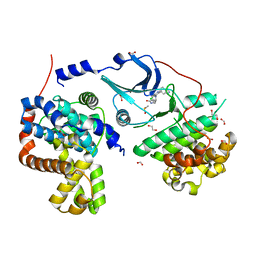 | | CDK8-CYCC IN COMPLEX WITH 8-[3-(3-Amino-1H-indazol-6-yl)-5-chloro- pyridine-4-yl]-2,8-diaza-spiro[4.5]decan-1-one | | Descriptor: | 1,2-ETHANEDIOL, 8-[3-(3-azanyl-2~{H}-indazol-6-yl)-5-chloranyl-pyridin-4-yl]-2,8-diazaspiro[4.5]decan-1-one, Cyclin-C, ... | | Authors: | Musil, D, Blagg, J, Mallinger, A. | | Deposit date: | 2015-12-20 | | Release date: | 2016-02-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Discovery of Potent, Selective, and Orally Bioavailable Small-Molecule Modulators of the Mediator Complex-Associated Kinases CDK8 and CDK19.
J.Med.Chem., 59, 2016
|
|
5HBE
 
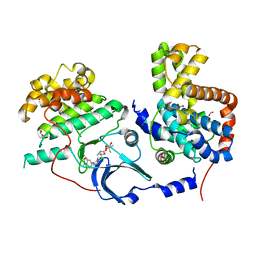 | | CDK8-CYCC IN COMPLEX WITH 8-[3-Chloro-5-(1-methyl-2,2-dioxo-2, 3-dihydro-1H-2l6-benzo[c]isothiazol-5-yl)-pyridin- 4-yl]-1-oxa-3,8-diaza-spiro[4.5]decan-2-one | | Descriptor: | 1,2-ETHANEDIOL, 8-[3-chloranyl-5-[1-methyl-2,2-bis(oxidanylidene)-3~{H}-2,1-benzothiazol-5-yl]pyridin-4-yl]-1-oxa-3,8-diazaspiro[4.5]decan-2-one, Cyclin-C, ... | | Authors: | Musil, D, Blagg, J, Mallinger, A. | | Deposit date: | 2015-12-31 | | Release date: | 2016-02-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Discovery of Potent, Selective, and Orally Bioavailable Small-Molecule Modulators of the Mediator Complex-Associated Kinases CDK8 and CDK19.
J.Med.Chem., 59, 2016
|
|
5I5Z
 
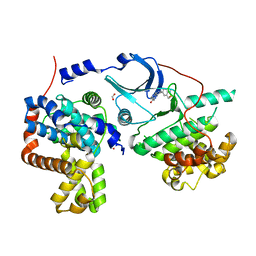 | | CDK8-CYCC IN COMPLEX WITH 8-(1-Methyl-2,2-dioxo-2,3-dihydro-1H-2l6-benzo[c]isothiazol-5-yl)-[1,6]naphthyridine-2-carboxylic acid methylamide | | Descriptor: | Cyclin-C, Cyclin-dependent kinase 8, FORMIC ACID, ... | | Authors: | Musil, D, Blagg, J, Mallinger, A. | | Deposit date: | 2016-02-15 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | 2,8-Disubstituted-1,6-Naphthyridines and 4,6-Disubstituted-Isoquinolines with Potent, Selective Affinity for CDK8/19.
Acs Med.Chem.Lett., 7, 2016
|
|
5HBJ
 
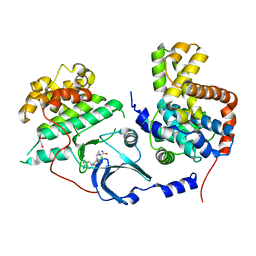 | | CDK8-CYCC IN COMPLEX WITH 8-[2-Amino-3-chloro-5-(1-methyl-1H-indazol-5-yl)-pyridin-4-yl]-2,8-diaza-spiro[4.5]decan-1-one | | Descriptor: | 1,2-ETHANEDIOL, 8-[2-azanyl-3-chloranyl-5-(1-methylindazol-5-yl)pyridin-4-yl]-2,8-diazaspiro[4.5]decan-1-one, Cyclin-C, ... | | Authors: | Musil, D, Blagg, J, Mallinger, A. | | Deposit date: | 2015-12-31 | | Release date: | 2016-02-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Discovery of Potent, Selective, and Orally Bioavailable Small-Molecule Modulators of the Mediator Complex-Associated Kinases CDK8 and CDK19.
J.Med.Chem., 59, 2016
|
|
5HBH
 
 | | CDK8-CYCC IN COMPLEX WITH 5-{5-Chloro-4-[1-(2-methoxy-ethyl)-1,8-diaza-spiro[4.5]dec-8-yl]-pyridin-3-yl}-1-methyl-1,3-dihydro-benzo[c]isothiazole 2,2-dioxide | | Descriptor: | 1,2-ETHANEDIOL, 5-[5-chloranyl-4-[1-(2-methoxyethyl)-1,8-diazaspiro[4.5]decan-8-yl]pyridin-3-yl]-1-methyl-3~{H}-2,1-benzothiazole 2,2-dioxide, Cyclin-C, ... | | Authors: | Musil, D, Blagg, J, Mallinger, A. | | Deposit date: | 2015-12-31 | | Release date: | 2016-02-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of Potent, Selective, and Orally Bioavailable Small-Molecule Modulators of the Mediator Complex-Associated Kinases CDK8 and CDK19.
J.Med.Chem., 59, 2016
|
|
9EUV
 
 | |
9EUW
 
 | |
3TZ8
 
 | | Kinase domain of cSrc in complex with RL104 | | Descriptor: | N-(4-{[4-({[1-(3-aminophenyl)-3-tert-butyl-1H-pyrazol-5-yl]carbamoyl}amino)phenyl]amino}quinazolin-6-yl)-3-(4-methylpiperazin-1-yl)propanamide, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Gruetter, C, Richters, A, Rauh, D. | | Deposit date: | 2011-09-27 | | Release date: | 2012-10-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Overcoming Gatekeeper Mutations in cSrc and Abl by Hybrid Compound Design
To be Published
|
|
3TZ9
 
 | | Kinase domain of cSrc in complex with RL130 | | Descriptor: | 1-[1-(3-aminophenyl)-3-tert-butyl-1H-pyrazol-5-yl]-3-[4-(quinazolin-4-ylamino)phenyl]urea, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Gruetter, C, Richters, A, Rauh, D. | | Deposit date: | 2011-09-27 | | Release date: | 2012-10-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Overcoming Gatekeeper Mutations in cSrc and Abl by Hybrid Compound Design
To be Published
|
|
3TZ7
 
 | | Kinase domain of cSrc in complex with RL103 | | Descriptor: | N-(4-{[4-({[1-(3-aminophenyl)-3-tert-butyl-1H-pyrazol-5-yl]carbamoyl}amino)phenyl]amino}quinazolin-6-yl)-4-(dimethylamino)butanamide, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Gruetter, C, Richters, A, Rauh, D. | | Deposit date: | 2011-09-27 | | Release date: | 2012-10-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Overcoming Gatekeeper Mutations in cSrc and Abl by Hybrid Compound Design
To be Published
|
|
7ZJQ
 
 | |
