1BXL
 
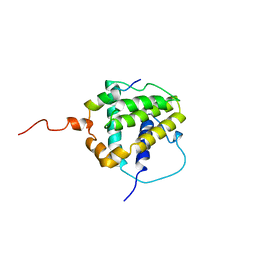 | | STRUCTURE OF BCL-XL/BAK PEPTIDE COMPLEX, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | BAK PEPTIDE, BCL-XL | | Authors: | Sattler, M, Liang, H, Nettesheim, D, Meadows, R.P, Harlan, J.E, Eberstadt, M, Yoon, H, Shuker, S.B, Chang, B.S, Minn, A.J, Thompson, C.B, Fesik, S.W. | | Deposit date: | 1996-10-16 | | Release date: | 1997-10-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of Bcl-xL-Bak peptide complex: recognition between regulators of apoptosis.
Science, 275, 1997
|
|
2MJN
 
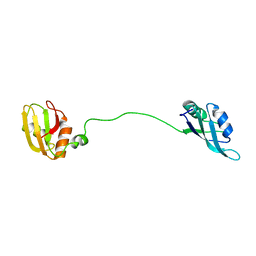 | |
8P25
 
 | | Solution structure of a chimeric U2AF2 RRM2 / FUBP1 N-Box | | Descriptor: | Splicing factor U2AF 65 kDa subunit,Far upstream element-binding protein 1 | | Authors: | Hipp, C, Sattler, M. | | Deposit date: | 2023-05-14 | | Release date: | 2023-07-26 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | FUBP1 is a general splicing factor facilitating 3' splice site recognition and splicing of long introns.
Mol.Cell, 83, 2023
|
|
4UQT
 
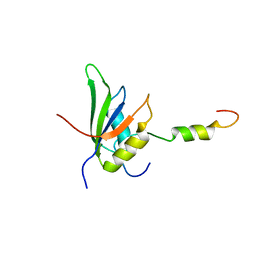 | | RRM-peptide structure in RES complex | | Descriptor: | PRE-MRNA-SPLICING FACTOR CWC26, U2 SNRNP COMPONENT IST3 | | Authors: | Tripsianes, K, Friberg, A, Barrandon, C, Seraphin, B, Sattler, M. | | Deposit date: | 2014-06-25 | | Release date: | 2014-09-03 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | A Novel Protein-Protein Interaction in the Res (Retention and Splicing) Complex.
J.Biol.Chem., 289, 2014
|
|
5JU7
 
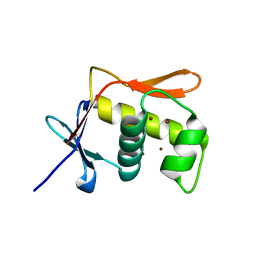 | | DNA BINDING DOMAIN OF E.COLI CADC | | Descriptor: | Transcriptional activator CadC, ZINC ION | | Authors: | Janowski, R, Schlundt, A, Sattler, M, Niessing, D. | | Deposit date: | 2016-05-10 | | Release date: | 2017-04-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure-function analysis of the DNA-binding domain of a transmembrane transcriptional activator.
Sci Rep, 7, 2017
|
|
8S3G
 
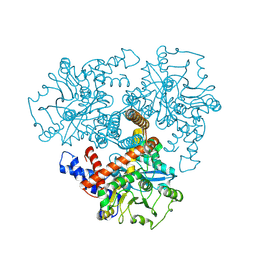 | |
6I68
 
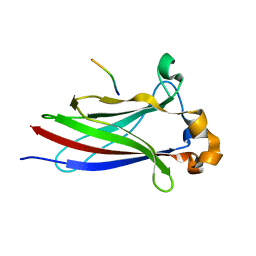 | |
7NDX
 
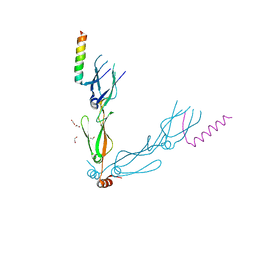 | | Crystal structure of the human HSP40 DNAJB1-CTDs in complex with a peptide of NudC | | Descriptor: | 1,2-ETHANEDIOL, DnaJ homolog subfamily B member 1, Nuclear migration protein nudC | | Authors: | Delhommel, F, Zak, K.M, Popowicz, G.M, Sattler, M. | | Deposit date: | 2021-02-02 | | Release date: | 2022-01-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.541 Å) | | Cite: | NudC guides client transfer between the Hsp40/70 and Hsp90 chaperone systems.
Mol.Cell, 82, 2022
|
|
8B8S
 
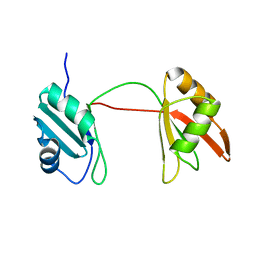 | | Solution structure of tandem RRM1 and RRM2 domains of yeast NPL3 | | Descriptor: | Serine/arginine (SR)-type shuttling mRNA binding protein NPL3 | | Authors: | Kachariya, N, Sattler, M, Keil, P, Strasser, K. | | Deposit date: | 2022-10-04 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | Npl3 functions in mRNP assembly by recruitment of mRNP components to the transcription site and their transfer onto the mRNA.
Nucleic Acids Res., 51, 2023
|
|
2JW6
 
 | | Solution structure of the DEAF1 MYND domain | | Descriptor: | Deformed epidermal autoregulatory factor 1 homolog, ZINC ION | | Authors: | Spadaccini, R, Perrin, H, Bottomley, M, Ansieu, S, Sattler, M. | | Deposit date: | 2007-10-08 | | Release date: | 2007-12-25 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Retraction notice to "Structure and functional analysis of the MYND domain" [J. Mol. Biol. (2006) 358, 498-508].
J.Mol.Biol., 376, 2008
|
|
3DXB
 
 | | Structure of the UHM domain of Puf60 fused to thioredoxin | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, thioredoxin N-terminally fused to Puf60(UHM) | | Authors: | Corsini, L, Hothorn, M, Scheffzek, K, Stier, G, Sattler, M. | | Deposit date: | 2008-07-24 | | Release date: | 2008-10-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Dimerization and Protein Binding Specificity of the U2AF Homology Motif of the Splicing Factor Puf60.
J.Biol.Chem., 284, 2009
|
|
9GAG
 
 | |
2JXC
 
 | | Structure of the EPS15-EH2 Stonin2 Complex | | Descriptor: | CALCIUM ION, Epidermal growth factor receptor substrate 15, Stonin-2 | | Authors: | Rumpf, J, Simon, B, Jung, N, Maritzen, T, Haucke, V, Sattler, M, Groemping, Y. | | Deposit date: | 2007-11-12 | | Release date: | 2008-01-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of the Eps15-stonin2 complex provides a molecular explanation for EH-domain ligand specificity.
Embo J., 27, 2008
|
|
9I8B
 
 | | NMR-based model of an A-form 20mer dsRNA | | Descriptor: | RNA (5'-R(*AP*CP*UP*GP*GP*AP*CP*AP*AP*AP*UP*AP*CP*UP*CP*CP*GP*AP*GP*G)-3'), RNA (5'-R(*CP*CP*UP*CP*GP*GP*AP*GP*UP*AP*UP*UP*UP*GP*UP*CP*CP*AP*GP*U)-3') | | Authors: | Mueller-Hermes, C, Sattler, M. | | Deposit date: | 2025-02-04 | | Release date: | 2025-03-12 | | Method: | SOLUTION NMR | | Cite: | NMR-based model of an A-form 20mer dsRNA
To Be Published
|
|
1FHO
 
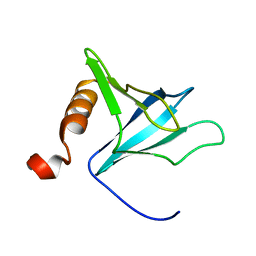 | | Solution Structure of the PH Domain from the C. Elegans Muscle Protein UNC-89 | | Descriptor: | UNC-89 | | Authors: | Blomberg, N, Baraldi, E, Sattler, M, Saraste, M, Nilges, M. | | Deposit date: | 2000-08-02 | | Release date: | 2000-10-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of a PH domain from the C. elegans muscle protein UNC-89 suggests a novel function.
Structure Fold.Des., 8, 2000
|
|
1D8B
 
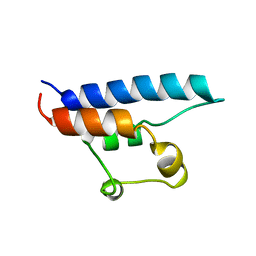 | | NMR STRUCTURE OF THE HRDC DOMAIN FROM SACCHAROMYCES CEREVISIAE RECQ HELICASE | | Descriptor: | SGS1 RECQ HELICASE | | Authors: | Liu, Z, Macias, M.J, Bottomley, M.J, Stier, G, Linge, J.P, Nilges, M, Bork, P, Sattler, M. | | Deposit date: | 1999-10-21 | | Release date: | 2000-01-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The three-dimensional structure of the HRDC domain and implications for the Werner and Bloom syndrome proteins.
Structure Fold.Des., 7, 1999
|
|
4EL6
 
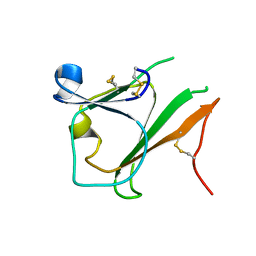 | | Crystal structure of IPSE/alpha-1 from Schistosoma mansoni eggs | | Descriptor: | IL-4-inducing protein | | Authors: | Mayerhofer, H, Meyer, H, Tripsianes, K, Barths, D, Blindow, S, Bade, S, Madl, T, Frey, A, Haas, H, Sattler, M, Schramm, G, Mueller-Dieckmann, J. | | Deposit date: | 2012-04-10 | | Release date: | 2013-04-10 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | A Crystallin Fold in the Interleukin-4-inducing Principle of Schistosoma mansoni Eggs (IPSE/ alpha-1) Mediates IgE Binding for Antigen-independent Basophil Activation.
J.Biol.Chem., 290, 2015
|
|
3NC1
 
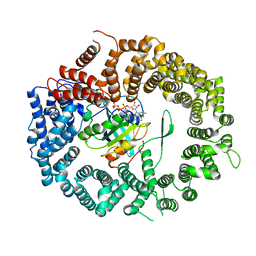 | | Crystal structure of the CRM1-RanGTP complex | | Descriptor: | Exportin-1, GTP-binding nuclear protein Ran, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Guttler, T, Madl, T, Neumann, P, Deichsel, D, Corsini, L, Monecke, T, Ficner, R, Sattler, M, Gorlich, D. | | Deposit date: | 2010-06-04 | | Release date: | 2010-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | NES consensus redefined by structures of PKI-type and Rev-type nuclear export signals bound to CRM1.
Nat.Struct.Mol.Biol., 17, 2010
|
|
5AGQ
 
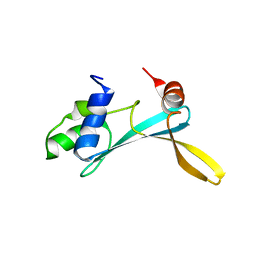 | | Solution structure of the TAM domain of human TIP5 BAZ2A involved in epigenetic regulation of rRNA genes | | Descriptor: | BROMODOMAIN ADJACENT TO ZINC FINGER DOMAIN PROTEIN 2A | | Authors: | Anosova, I, Melnik, S, Tripsianes, K, Kateb, F, Grummt, I, Sattler, M. | | Deposit date: | 2015-02-03 | | Release date: | 2015-05-06 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | A Novel RNA Binding Surface of the Tam Domain of Tip5/Baz2A Mediates Epigenetic Regulation of Rrna Genes.
Nucleic Acids Res., 43, 2015
|
|
8POI
 
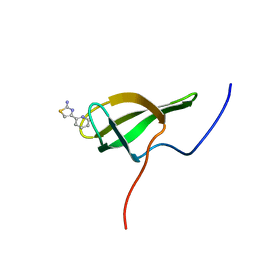 | |
1T2S
 
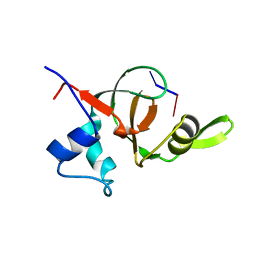 | | Structural basis for 3' end recognition of nucleic acids by the Drosophila Argonaute 2 PAZ domain | | Descriptor: | 5'-D(*CP*TP*CP*AP*C)-3', Argonaute 2 | | Authors: | Lingel, A, Simon, B, Izaurralde, E, Sattler, M. | | Deposit date: | 2004-04-22 | | Release date: | 2004-06-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Nucleic acid 3'-end recognition by the Argonaute2 PAZ domain.
Nat.Struct.Mol.Biol., 11, 2004
|
|
1T2R
 
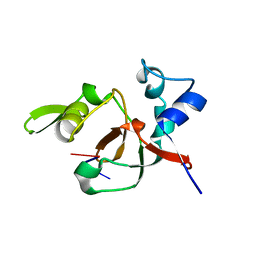 | | Structural basis for 3' end recognition of nucleic acids by the Drosophila Argonaute 2 PAZ domain | | Descriptor: | 5'-R(*CP*UP*CP*AP*C)-3', Argonaute 2 | | Authors: | Lingel, A, Simon, B, Izaurralde, E, Sattler, M. | | Deposit date: | 2004-04-22 | | Release date: | 2004-06-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Nucleic acid 3'-end recognition by the Argonaute2 PAZ domain.
Nat.Struct.Mol.Biol., 11, 2004
|
|
8PXX
 
 | |
8PXW
 
 | |
1MHN
 
 | |
