8VM1
 
 | |
8TM0
 
 | |
8TLZ
 
 | |
8TM2
 
 | |
1M5K
 
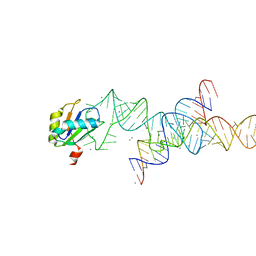 | |
1SKN
 
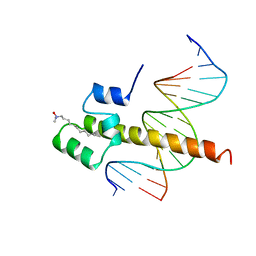 | | THE BINDING DOMAIN OF SKN-1 IN COMPLEX WITH DNA: A NEW DNA-BINDING MOTIF | | Descriptor: | DNA (5'-D(*CP*AP*GP*GP*GP*AP*TP*GP*AP*CP*AP*TP*TP*GP*T)-3'), DNA (5'-D(*TP*GP*AP*CP*AP*AP*TP*GP*TP*CP*AP*TP*CP*CP*C)-3'), DNA-BINDING DOMAIN OF SKN-1, ... | | Authors: | Rupert, P.B, Daughdrill, G.W, Bowerman, B, Matthews, B.W. | | Deposit date: | 1998-03-30 | | Release date: | 1998-06-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A new DNA-binding motif in the Skn-1 binding domain-DNA complex.
Nat.Struct.Biol., 5, 1998
|
|
1M5V
 
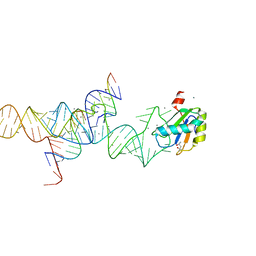 | | Transition State Stabilization by a Catalytic RNA | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, RNA HAIRPIN RIBOZYME, ... | | Authors: | Rupert, P.B, Massey, A.P, Sigurdsson, S.T, Ferre-D'Amare, A.R. | | Deposit date: | 2002-07-09 | | Release date: | 2002-10-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Transition state stabilization by a catalytic RNA
Science, 298, 2002
|
|
1M5O
 
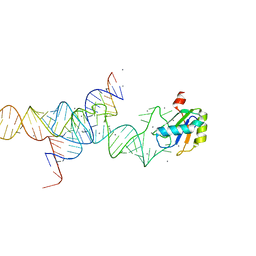 | | Transition State Stabilization by a Catalytic RNA | | Descriptor: | CALCIUM ION, RNA SUBSTRATE, RNA HAIRPIN RIBOZYME, ... | | Authors: | Rupert, P.B, Massey, A.P, Sigurdsson, S.T, Ferre-D'Amare, A.R. | | Deposit date: | 2002-07-09 | | Release date: | 2002-10-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Transition state stabilization by a catalytic RNA
Science, 298, 2002
|
|
1M5P
 
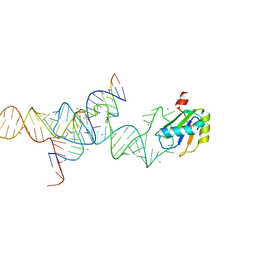 | | Transition State Stabilization by a Catalytic RNA | | Descriptor: | CALCIUM ION, RNA HAIRPIN RIBOZYME, RNA INHIBITOR SUBSTRATE, ... | | Authors: | Rupert, P.B, Massey, A, Sigurdsson, S.T, Ferre-D'Amare, A.R. | | Deposit date: | 2002-07-09 | | Release date: | 2002-10-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Transition state stabilization by a catalytic RNA
Science, 298, 2002
|
|
1D1L
 
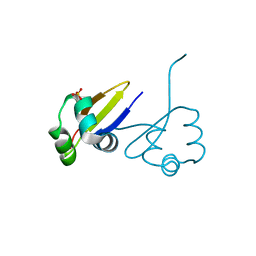 | | CRYSTAL STRUCTURE OF CRO-F58W MUTANT | | Descriptor: | LAMBDA CRO REPRESSOR, SULFATE ION | | Authors: | Rupert, P.B, Mollah, A.K, Mossing, M.C, Matthews, B.W. | | Deposit date: | 1999-09-17 | | Release date: | 1999-10-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structural basis for enhanced stability and reduced DNA binding seen in engineered second-generation Cro monomers and dimers.
J.Mol.Biol., 296, 2000
|
|
1D1M
 
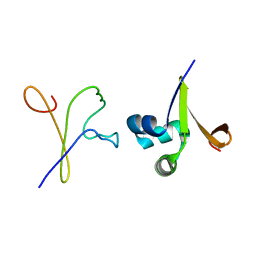 | |
1NU4
 
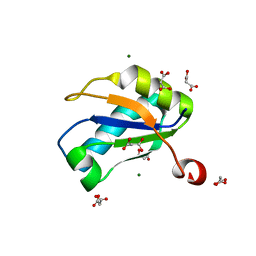 | | U1A RNA binding domain at 1.8 angstrom resolution reveals a pre-organized C-terminal helix | | Descriptor: | MAGNESIUM ION, MALONIC ACID, U1A RNA binding domain | | Authors: | Rupert, P.B, Xiao, H, Ferre-D'Amare, A.R. | | Deposit date: | 2003-01-30 | | Release date: | 2003-02-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | U1A RNA-binding domain at 1.8 A resolution.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
7SJQ
 
 | |
6O5D
 
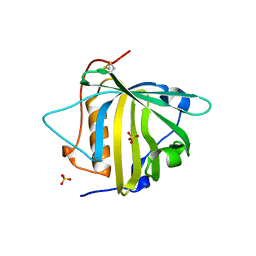 | | PYOCHELIN | | Descriptor: | GLYCEROL, Neutrophil gelatinase-associated lipocalin, SULFATE ION | | Authors: | Rupert, P.B, Strong, R.K, Clifton, M.C, Edwards, T.E, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2019-03-01 | | Release date: | 2019-09-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Parsing the functional specificity of Siderocalin/Lipocalin 2/NGAL for siderophores and related small-molecule ligands.
J Struct Biol X, 2, 2019
|
|
6OKE
 
 | |
5JR8
 
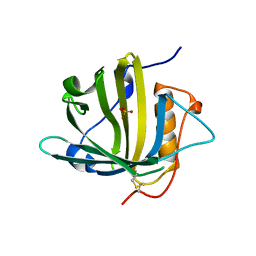 | | Disposal of Iron by a Mutant form of Siderocalin NGAL | | Descriptor: | GLYCEROL, Neutrophil gelatinase-associated lipocalin, PHOSPHATE ION | | Authors: | Rupert, P.B, Strong, R.K, Barasch, J, Hollman, M, Deng, R, Hod, E.A, Abergel, R, Allred, B, Xu, K, Darrah, S, Tekabe, Y, Perlstein, A, Bruck, E, Stauber, J, Corbin, K, Buchen, C, Slavkovich, V, Graziano, J, Spitalnik, S, Qiu, A. | | Deposit date: | 2016-05-05 | | Release date: | 2016-09-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Disposal of iron by a mutant form of lipocalin 2.
Nat Commun, 7, 2016
|
|
5KID
 
 | | Tightening the Recognition of Tetravalent Zr and Th Complexes by the Siderophore-Binding Mammalian Protein Siderocalin for Theranostic Applications | | Descriptor: | CHLORIDE ION, GLYCEROL, N,N'-(butane-1,4-diyl)bis(N-{3-[(2,3-dihydroxybenzene-1-carbonyl)amino]propyl}-2,3-dihydroxybenzamide), ... | | Authors: | Rupert, P.B, Strong, R.K. | | Deposit date: | 2016-06-16 | | Release date: | 2017-04-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Engineered Recognition of Tetravalent Zirconium and Thorium by Chelator-Protein Systems: Toward Flexible Radiotherapy and Imaging Platforms.
Inorg Chem, 55, 2016
|
|
5KHP
 
 | | Tightening the Recognition of Tetravalent Zr and Th Complexes by the Siderophore-Binding Mammalian Protein Siderocalin for Theranostic Applications | | Descriptor: | GLYCEROL, N,N'-(butane-1,4-diyl)bis(N-{3-[(2,3-dihydroxybenzene-1-carbonyl)amino]propyl}-2,3-dihydroxybenzamide), Neutrophil gelatinase-associated lipocalin, ... | | Authors: | Rupert, P.B, Strong, R.K. | | Deposit date: | 2016-06-15 | | Release date: | 2017-04-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Engineered Recognition of Tetravalent Zirconium and Thorium by Chelator-Protein Systems: Toward Flexible Radiotherapy and Imaging Platforms.
Inorg Chem, 55, 2016
|
|
5KIC
 
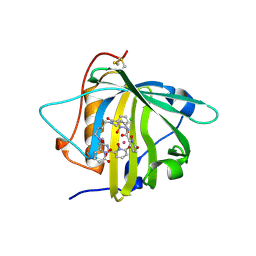 | | Long-sought stabilization of berkelium(IV) in solution: An anomaly within the heavy actinide series | | Descriptor: | CALIFORNIUM ION, CHLORIDE ION, N,N'-butane-1,4-diylbis[1-hydroxy-N-(3-{[(1-hydroxy-6-oxo-1,6-dihydropyridin-2-yl)carbonyl]amino}propyl)-6-oxo-1,6-dihydropyridine-2-carboxamide], ... | | Authors: | Rupert, P.B, Strong, R.K. | | Deposit date: | 2016-06-16 | | Release date: | 2017-04-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Chelation and stabilization of berkelium in oxidation state +IV.
Nat Chem, 9, 2017
|
|
4LLV
 
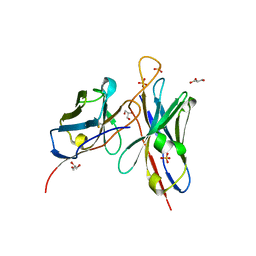 | | The structure of the unbound form of anti-HIV antibody 4E10 Fv | | Descriptor: | 4E10 Fv heavy chain, 4E10 Fv light chain, GLYCEROL, ... | | Authors: | Finton, K.A.K, Rupert, P.B, Strong, R.K. | | Deposit date: | 2013-07-09 | | Release date: | 2013-10-16 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Autoreactivity and Exceptional CDR Plasticity (but Not Unusual Polyspecificity) Hinder Elicitation of the Anti-HIV Antibody 4E10.
Plos Pathog., 9, 2013
|
|
6OKD
 
 | | Crystal Structure of human transferrin receptor in complex with a cystine-dense peptide | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Finton, K.A.K, Rupert, P.B, Strong, R.K. | | Deposit date: | 2019-04-12 | | Release date: | 2020-04-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A TfR-Binding Cystine-Dense Peptide Promotes Blood-Brain Barrier Penetration of Bioactive Molecules.
J.Mol.Biol., 432, 2020
|
|
4ZFX
 
 | | Siderocalin-mediated recognition and cellular uptake of actinides | | Descriptor: | N,N',N''-[(3S,7S,11S)-2,6,10-trioxo-1,5,9-trioxacyclododecane-3,7,11-triyl]tris(2,3-dihydroxybenzamide), Neutrophil gelatinase-associated lipocalin, SULFATE ION, ... | | Authors: | Allred, B.E, Rupert, P.B, Gauny, S.S, An, D.D, Ralston, C.Y, Sturzbecher-Hoehne, M, Strong, R.K, Abergel, R.J. | | Deposit date: | 2015-04-22 | | Release date: | 2015-08-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Siderocalin-mediated recognition, sensitization, and cellular uptake of actinides.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4ZHD
 
 | | Siderocalin-mediated recognition and cellular uptake of actinides | | Descriptor: | GLYCEROL, Neutrophil gelatinase-associated lipocalin, PLUTONIUM ION, ... | | Authors: | Allred, B.E, Rupert, P.B, Gauny, S.S, An, D.D, Ralston, C.Y, Sturzbecher-Hoehne, M, Strong, R.K, Abergel, R.J. | | Deposit date: | 2015-04-24 | | Release date: | 2015-08-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Siderocalin-mediated recognition, sensitization, and cellular uptake of actinides.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4ZHG
 
 | | Siderocalin-mediated recognition and cellular uptake of actinides | | Descriptor: | AMERICIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Allred, B.E, Rupert, P.B, Gauny, S.S, An, D.D, Ralston, C.Y, Sturzbecher-Hoehne, M, Strong, R.K, Abergel, R.J. | | Deposit date: | 2015-04-24 | | Release date: | 2015-08-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Siderocalin-mediated recognition, sensitization, and cellular uptake of actinides.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4ZHH
 
 | | Siderocalin-mediated recognition and cellular uptake of actinides | | Descriptor: | CHLORIDE ION, GLYCEROL, N,N'-butane-1,4-diylbis[1-hydroxy-N-(3-{[(1-hydroxy-6-oxo-1,6-dihydropyridin-2-yl)carbonyl]amino}propyl)-6-oxo-1,6-dihydropyridine-2-carboxamide], ... | | Authors: | Allred, B.E, Rupert, P.B, Gauny, S.S, An, D.D, Ralston, C.Y, Sturzbecher-Hoehne, M, Strong, R.K, Abergel, R.J. | | Deposit date: | 2015-04-24 | | Release date: | 2015-08-05 | | Last modified: | 2015-09-02 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Siderocalin-mediated recognition, sensitization, and cellular uptake of actinides.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
