2A2T
 
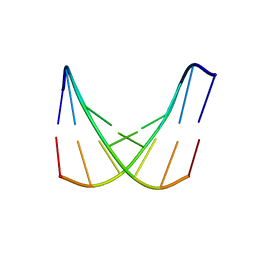 | | crystal structure of d(AAATATTT) | | Descriptor: | 5'-D(*AP*AP*AP*TP*AP*TP*TP*T)-3', CACODYLATE ION, MANGANESE (II) ION | | Authors: | Valls, N, Richter, M, Subirana, J.A. | | Deposit date: | 2005-06-23 | | Release date: | 2005-11-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of a DNA duplex with all-AT base pairs.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
8ROY
 
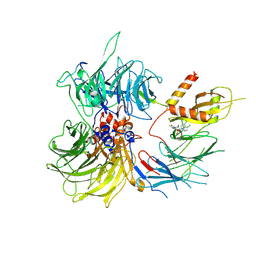 | | Structure of the human DDB1-DDA1-DCAF15 E3 ubiquitin ligase bound to compound furan 24 | | Descriptor: | 1-[5-[[3,4-bis(chloranyl)-1~{H}-indol-7-yl]sulfamoyl]-3-methyl-furan-2-yl]carbonyl-~{N}-methyl-piperidine-4-carboxamide, DDB1- and CUL4-associated factor 15, DET1- and DDB1-associated protein 1, ... | | Authors: | Shilliday, F, Lucas, S.C.C, Richter, M, Michaelides, I.N, Fusani, L. | | Deposit date: | 2024-01-12 | | Release date: | 2024-04-03 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Optimization of Potent Ligands for the E3 Ligase DCAF15 and Evaluation of Their Use in Heterobifunctional Degraders.
J.Med.Chem., 67, 2024
|
|
8ROX
 
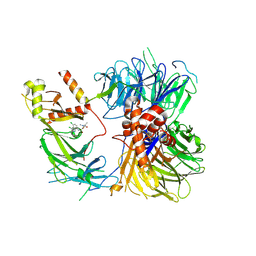 | | Structure of the human DDB1-DDA1-DCAF15 E3 ubiquitin ligase bound to compound furan 12 | | Descriptor: | 5-[[3,4-bis(chloranyl)-1~{H}-indol-7-yl]sulfamoyl]-~{N},~{N},3-trimethyl-furan-2-carboxamide;ethane, DDB1- and CUL4-associated factor 15, DET1- and DDB1-associated protein 1, ... | | Authors: | Shilliday, F, Lucas, S.C.C, Richter, M, Michaelides, I.N, Fusani, L. | | Deposit date: | 2024-01-12 | | Release date: | 2024-04-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Optimization of Potent Ligands for the E3 Ligase DCAF15 and Evaluation of Their Use in Heterobifunctional Degraders.
J.Med.Chem., 67, 2024
|
|
7KZ7
 
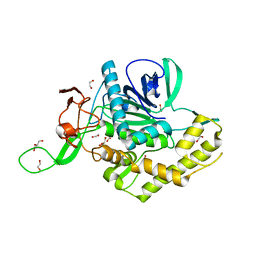 | | Crystals Structure of the Mutated Protease Domain of Botulinum Neurotoxin X (X4130B1). | | Descriptor: | 1,2-ETHANEDIOL, Botulinum neurotoxin type X, GLYCEROL, ... | | Authors: | Blum, T.R, Liu, H, Packer, M.S, Xiong, X, Lee, P.G, Zhang, S, Richter, M, Minasov, G, Satchell, K.J.F, Dong, M, Liu, D.R, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-12-10 | | Release date: | 2020-12-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Phage-assisted evolution of botulinum neurotoxin proteases with reprogrammed specificity.
Science, 371, 2021
|
|
4UZI
 
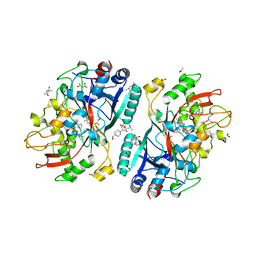 | | Crystal Structure of AauDyP Complexed with Imidazole | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Strittmatter, E, Liers, C, Ullrich, R, Hofrichter, M, Piontek, K, Plattner, D.A. | | Deposit date: | 2014-09-05 | | Release date: | 2015-01-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Toolbox of Auricularia Auricula-Judae Dye-Decolorizing Peroxidase - Identification of Three New Potential Substrate-Interaction Sites.
Arch.Biochem.Biophys., 574, 2015
|
|
2YP1
 
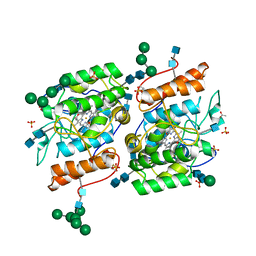 | | Crystallization of a 45 kDa peroxygenase- peroxidase from the mushroom Agrocybe aegerita and structure determination by SAD utilizing only the haem iron | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Piontek, K, Strittmatter, E, Ullrich, R, Plattner, D.A, Hofrichter, M. | | Deposit date: | 2012-10-29 | | Release date: | 2013-10-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structural Basis of Substrate Conversion in a New Aromatic Peroxygenase: P450 Functionality with Benefits
J.Biol.Chem., 288, 2013
|
|
2YOR
 
 | | Crystallization of a 45 kDa peroxygenase- peroxidase from the mushroom Agrocybe aegerita and structure determination by SAD utilizing only the haem iron | | Descriptor: | 1H-imidazol-5-ylmethanol, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Piontek, K, Strittmatter, E, Ullrich, R, Plattner, D.A, Hofrichter, M. | | Deposit date: | 2012-10-26 | | Release date: | 2013-10-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structural Basis of Substrate Conversion in a New Aromatic Peroxygenase: P450 Functionality with Benefits
J.Biol.Chem., 288, 2013
|
|
4AU9
 
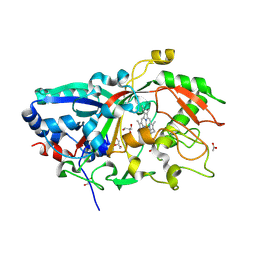 | | Crystal Structure of a Fungal DyP-Type Peroxidase from Auricularia auricula-judae | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Strittmatter, E, Pecyna, M, Ullrich, R, Hofrichter, M, Plattner, D.A, Liers, C, Piontek, K. | | Deposit date: | 2012-05-14 | | Release date: | 2012-12-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | First Crystal Structure of a Fungal High-Redox Potential Dye-Decolorizing Peroxidase: Substrate Interaction Sites and Long-Range Electron Transfer.
J.Biol.Chem., 288, 2013
|
|
6F6O
 
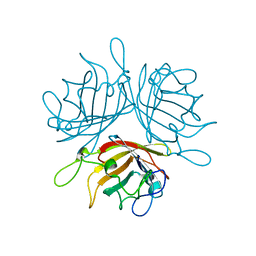 | |
4WYJ
 
 | | Adenovirus 3 head domain mutant V239D | | Descriptor: | Fiber protein, SULFATE ION | | Authors: | Lieber, A, Zubieta, C, Fender, P. | | Deposit date: | 2014-11-17 | | Release date: | 2015-07-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Preclinical safety and efficacy studies with an affinity-enhanced epithelial junction opener and PEGylated liposomal doxorubicin.
Mol Ther Methods Clin Dev, 2, 2015
|
|
4LIY
 
 | |
5ANY
 
 | | Electron cryo-microscopy of chikungunya virus in complex with neutralizing antibody Fab CHK265 | | Descriptor: | E1, E2, FAB, ... | | Authors: | Fox, J.M, Long, F, Edeling, M.A, Lin, H, Duijl-Richter, M, Fong, R.H, Kahle, K.M, Smit, J.M, Jin, J, Simmons, G, Doranz, B.J, Crowe, J.E, Fremont, D.H, Rossmann, M.G, Diamond, M.S. | | Deposit date: | 2015-09-08 | | Release date: | 2015-11-25 | | Last modified: | 2018-10-03 | | Method: | ELECTRON MICROSCOPY (16.9 Å) | | Cite: | Broadly Neutralizing Alphavirus Antibodies Bind an Epitope on E2 and Inhibit Entry and Egress.
Cell(Cambridge,Mass.), 163, 2015
|
|
8TQD
 
 | | NF-Kappa-B1 Bound with a Covalent Inhibitor | | Descriptor: | 1-(2-bromo-4-chlorophenyl)-N-{(3S)-1-[(E)-iminomethyl]pyrrolidin-3-yl}methanesulfonamide, Nuclear factor NF-kappa-B p105 subunit | | Authors: | Hilbert, B.J. | | Deposit date: | 2023-08-07 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | DrugMap: A quantitative pan-cancer analysis of cysteine ligandability.
Cell, 187, 2024
|
|
6CR0
 
 | | 1.55 A resolution structure of (S)-6-hydroxynicotine oxidase from Shinella HZN7 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, (S)-6-hydroxynicotine oxidase, ACETATE ION, ... | | Authors: | Deay III, D, Lovell, S, Battaile, K.P, Petillo, P, Richter, M. | | Deposit date: | 2018-03-16 | | Release date: | 2018-03-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.548 Å) | | Cite: | Improving the kinetic parameters of nicotine oxidizing enzymes by homologous structure comparison and rational design
Arch.Biochem.Biophys., 2022
|
|
6DE6
 
 | |
4ZDG
 
 | |
6Z82
 
 | |
6SK7
 
 | | Cryo-EM structure of rhinovirus-A89 | | Descriptor: | VP1 capsid protein, VP2 capsid protein, VP3 capsid protein, ... | | Authors: | Wald, J, Goessweiner-Mohr, N, Blaas, D, Pasin, M. | | Deposit date: | 2019-08-14 | | Release date: | 2019-09-04 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM structure of pleconaril-resistant rhinovirus-B5 complexed to the antiviral OBR-5-340 reveals unexpected binding site.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6SK5
 
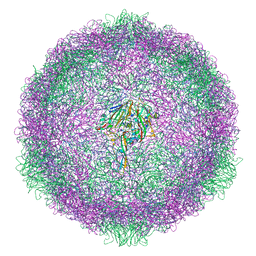 | | Cryo-EM structure of rhinovirus-B5 complexed to antiviral OBR-5-340 | | Descriptor: | 6-phenyl-~{N}3-[4-(trifluoromethyl)phenyl]-1~{H}-pyrazolo[3,4-d]pyrimidine-3,4-diamine, Rhinovirus B5 VP1, Rhinovirus B5 VP2, ... | | Authors: | Wald, J, Goessweiner-Mohr, N, Blaas, D, Pasin, M. | | Deposit date: | 2019-08-14 | | Release date: | 2019-09-04 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structure of pleconaril-resistant rhinovirus-B5 complexed to the antiviral OBR-5-340 reveals unexpected binding site.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6SK6
 
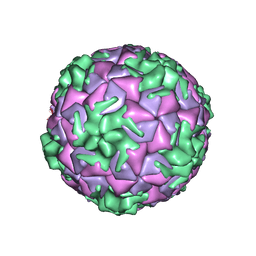 | | Cryo-EM structure of rhinovirus-B5 | | Descriptor: | Rhinovirus B5 VP1, Rhinovirus B5 VP2, Rhinovirus B5 VP3, ... | | Authors: | Wald, J, Goessweiner-Mohr, N, Blaas, D, Pasin, M. | | Deposit date: | 2019-08-14 | | Release date: | 2019-09-04 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of pleconaril-resistant rhinovirus-B5 complexed to the antiviral OBR-5-340 reveals unexpected binding site.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
4L4Q
 
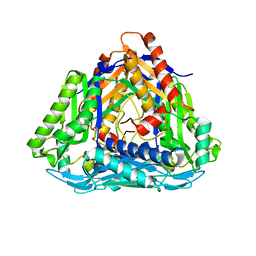 | | Methionine Adenosyltransferase | | Descriptor: | S-adenosylmethionine synthase | | Authors: | Schlesier, J, Siegrist, J, Gerhardt, S, Andexer, J.N, Einsle, O. | | Deposit date: | 2013-06-09 | | Release date: | 2014-03-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and functional characterisation of the methionine adenosyltransferase from Thermococcus kodakarensis.
Bmc Struct.Biol., 13, 2013
|
|
5LHM
 
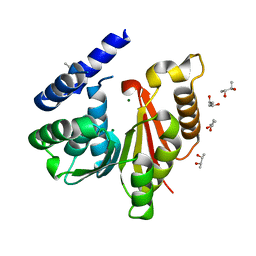 | | Crystal Structure of SafC from Myxococcus xanthus apo-Form | | Descriptor: | (R,R)-2,3-BUTANEDIOL, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Gerhardt, S, Netzer, J, Einsle, O. | | Deposit date: | 2016-07-12 | | Release date: | 2017-05-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Functional and structural characterisation of a bacterial O-methyltransferase and factors determining regioselectivity.
FEBS Lett., 591, 2017
|
|
5LOG
 
 | | Crystal Structure of SafC from Myxococcus xanthus bound to SAM | | Descriptor: | CHLORIDE ION, L-DOPAMINE, MAGNESIUM ION, ... | | Authors: | Gerhardt, S, Netzer, J, Einsle, O. | | Deposit date: | 2016-08-09 | | Release date: | 2017-06-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Functional and structural characterisation of a bacterial O-methyltransferase and factors determining regioselectivity.
FEBS Lett., 591, 2017
|
|
2GVD
 
 | |
2GVZ
 
 | |
