4RGF
 
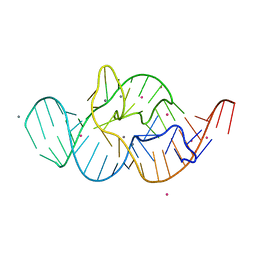 | | Crystal structure of the in-line aligned env22 twister ribozyme soaked with Mn2+ | | Descriptor: | MAGNESIUM ION, MANGANESE (II) ION, POTASSIUM ION, ... | | Authors: | Ren, A, Rajashankar, K.R, Simanshu, D, Patel, D. | | Deposit date: | 2014-09-30 | | Release date: | 2014-12-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.2008 Å) | | Cite: | In-line alignment and Mg(2+) coordination at the cleavage site of the env22 twister ribozyme.
Nat Commun, 5, 2014
|
|
4RGE
 
 | | Crystal structure of the in-line aligned env22 twister ribozyme | | Descriptor: | MAGNESIUM ION, env22 twister ribozyme | | Authors: | Ren, A, Rajashankar, K.R, Simanshu, D, Patel, D. | | Deposit date: | 2014-09-30 | | Release date: | 2014-12-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | In-line alignment and Mg(2+) coordination at the cleavage site of the env22 twister ribozyme.
Nat Commun, 5, 2014
|
|
5K7C
 
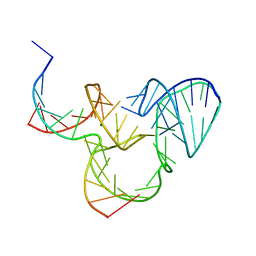 | | The native structure of native pistol ribozyme | | Descriptor: | DNA/RNA 11-MER, MAGNESIUM ION, RNA 47-MER | | Authors: | Ren, A, Patel, D. | | Deposit date: | 2016-05-26 | | Release date: | 2016-07-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Pistol ribozyme adopts a pseudoknot fold facilitating site-specific in-line cleavage.
Nat.Chem.Biol., 12, 2016
|
|
5K7D
 
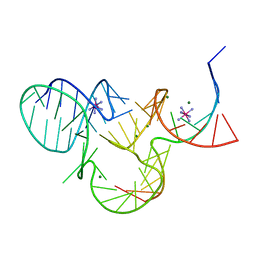 | | The structure of native pistol ribozyme, bound to Iridium | | Descriptor: | DNA/RNA 11-MER, IRIDIUM HEXAMMINE ION, MAGNESIUM ION, ... | | Authors: | Ren, A, Patel, D. | | Deposit date: | 2016-05-26 | | Release date: | 2016-07-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Pistol ribozyme adopts a pseudoknot fold facilitating site-specific in-line cleavage.
Nat.Chem.Biol., 12, 2016
|
|
5K7E
 
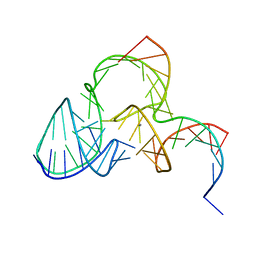 | | The structure of pistol ribozyme, soaked with Mn2+ | | Descriptor: | DNA/RNA 11-MER, MANGANESE (II) ION, RNA 47-MER | | Authors: | Ren, A, Patel, D. | | Deposit date: | 2016-05-26 | | Release date: | 2016-07-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.27 Å) | | Cite: | Pistol ribozyme adopts a pseudoknot fold facilitating site-specific in-line cleavage.
Nat.Chem.Biol., 12, 2016
|
|
5DDP
 
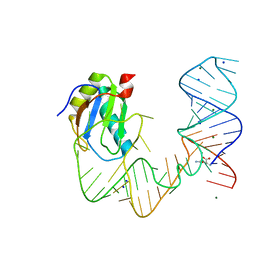 | | L-glutamine riboswitch bound with L-glutamine | | Descriptor: | GLUTAMINE, MAGNESIUM ION, RNA (61-MER), ... | | Authors: | Ren, A, Patel, D.J. | | Deposit date: | 2015-08-25 | | Release date: | 2015-12-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | Structural and Dynamic Basis for Low-Affinity, High-Selectivity Binding of L-Glutamine by the Glutamine Riboswitch.
Cell Rep, 13, 2015
|
|
5DDQ
 
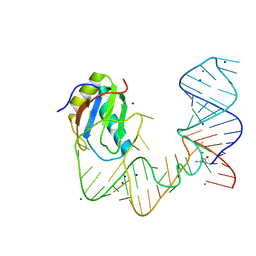 | | L-glutamine riboswitch bound with L-glutamine soaked with Mn2+ | | Descriptor: | GLUTAMINE, L-glutamine riboswitch RNA (61-MER), MAGNESIUM ION, ... | | Authors: | Ren, A, Patel, D.J. | | Deposit date: | 2015-08-25 | | Release date: | 2015-12-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and Dynamic Basis for Low-Affinity, High-Selectivity Binding of L-Glutamine by the Glutamine Riboswitch.
Cell Rep, 13, 2015
|
|
5DDR
 
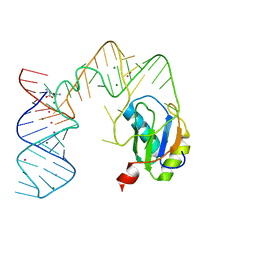 | | L-glutamine riboswitch bound with L-glutamine soaked with Cs+ | | Descriptor: | CESIUM ION, GLUTAMINE, L-glutamine riboswitch RNA (61-MER), ... | | Authors: | Ren, A, Patel, D.J. | | Deposit date: | 2015-08-25 | | Release date: | 2015-12-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.605 Å) | | Cite: | Structural and Dynamic Basis for Low-Affinity, High-Selectivity Binding of L-Glutamine by the Glutamine Riboswitch.
Cell Rep, 13, 2015
|
|
5DDO
 
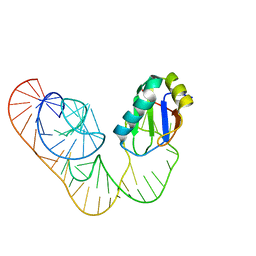 | |
4ZNP
 
 | | The structure of A pfI Riboswitch Bound to ZMP | | Descriptor: | AMINOIMIDAZOLE 4-CARBOXAMIDE RIBONUCLEOTIDE, MAGNESIUM ION, pfI Riboswitch | | Authors: | Ren, A, Patel, D.J, Rajashankar, R.K. | | Deposit date: | 2015-05-05 | | Release date: | 2015-08-26 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Global RNA Fold and Molecular Recognition for a pfl Riboswitch Bound to ZMP, a Master Regulator of One-Carbon Metabolism.
Structure, 23, 2015
|
|
4R96
 
 | | Structure of a Llama Glama Fab 48A2 against human cMet | | Descriptor: | Llama glama Fab 48A2 against human cMet H chain, Llama glama Fab 48A2 against human cMet L chain | | Authors: | Klarenbeek, A, El Mazouari, K, Desmyter, A, Blanchetot, C, Hultberg, A, Roovers, R.C, Cambillau, C, Spinelli, S, Del-Favero, J, Verrips, T, de Haard, H, Achour, I. | | Deposit date: | 2014-09-03 | | Release date: | 2015-06-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | Camelid Ig V genes reveal significant human homology not seen in therapeutic target genes, providing for a powerful therapeutic antibody platform.
MAbs, 7, 2015
|
|
4R90
 
 | | Anti CD70 Llama glama Fab 27B3 | | Descriptor: | Anti CD70 Llama glama Fab 27B3 Heavy chain, Anti CD70 Llama glama Fab 27B3 Light chain, CALCIUM ION, ... | | Authors: | Klarenbeek, A, El Mazouari, K, Desmyter, A, Blanchetot, C, Hultberg, A, Roovers, R.C, Cambillau, C, Spinelli, S, Del-Favero, J, Verrips, T, de Haard, H, Achour, I. | | Deposit date: | 2014-09-03 | | Release date: | 2015-06-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.746 Å) | | Cite: | Camelid Ig V genes reveal significant human homology not seen in therapeutic target genes, providing for a powerful therapeutic antibody platform.
MAbs, 7, 2015
|
|
5KAX
 
 | | The structure of CTR107 protein bound to RHODAMINE 6G | | Descriptor: | CTR107 protein, GLYCEROL, RHODAMINE 6G | | Authors: | Moreno, A, Wade, H. | | Deposit date: | 2016-06-02 | | Release date: | 2016-08-24 | | Last modified: | 2019-11-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Solution Binding and Structural Analyses Reveal Potential Multidrug Resistance Functions for SAV2435 and CTR107 and Other GyrI-like Proteins.
Biochemistry, 55, 2016
|
|
5KAU
 
 | | The structure of SAV2435 bound to RHODAMINE 6G | | Descriptor: | GLYCEROL, RHODAMINE 6G, SA2223 protein | | Authors: | Moreno, A, Wade, H. | | Deposit date: | 2016-06-02 | | Release date: | 2016-08-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Solution Binding and Structural Analyses Reveal Potential Multidrug Resistance Functions for SAV2435 and CTR107 and Other GyrI-like Proteins.
Biochemistry, 55, 2016
|
|
5KAW
 
 | |
5KAV
 
 | | The structure of SAV2435 | | Descriptor: | GLYCEROL, SA2223 protein | | Authors: | Moreno, A, Wade, H. | | Deposit date: | 2016-06-02 | | Release date: | 2016-08-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Solution Binding and Structural Analyses Reveal Potential Multidrug Resistance Functions for SAV2435 and CTR107 and Other GyrI-like Proteins.
Biochemistry, 55, 2016
|
|
5KAT
 
 | | The structure of SAV2435 bound to TETRAPHENYLPHOSPHONIUM | | Descriptor: | GLYCEROL, SA2223 protein, TETRAPHENYLPHOSPHONIUM | | Authors: | Moreno, A, Wade, H. | | Deposit date: | 2016-06-02 | | Release date: | 2016-08-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Solution Binding and Structural Analyses Reveal Potential Multidrug Resistance Functions for SAV2435 and CTR107 and Other GyrI-like Proteins.
Biochemistry, 55, 2016
|
|
5KCB
 
 | | The structure of SAV2435 bound to ethidium bromide | | Descriptor: | ETHIDIUM, SA2223 protein, SULFATE ION | | Authors: | Moreno, A, Wade, H. | | Deposit date: | 2016-06-06 | | Release date: | 2016-08-24 | | Last modified: | 2019-11-27 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Solution Binding and Structural Analyses Reveal Potential Multidrug Resistance Functions for SAV2435 and CTR107 and Other GyrI-like Proteins.
Biochemistry, 55, 2016
|
|
4O9H
 
 | | Structure of Interleukin-6 in complex with a Camelid Fab fragment | | Descriptor: | Heavy Chain of the Camelid Fab fragment 61H7, Interleukin-6, Light Chain of the Camelid Fab fragment 61H7 | | Authors: | Klarenbeek, A, Blanchetot, C, Schragel, G, Sadi, A.S, Ongenae, N, Hemrika, W, Wijdenes, J, Spinelli, S, Desmyter, A, Cambillau, C, Hultberg, A, Kretz-rommel, A, Dreier, T, De haard, H.J.W, Roovers, R.C. | | Deposit date: | 2014-01-02 | | Release date: | 2015-04-15 | | Last modified: | 2015-04-29 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Combining residues of naturally-occurring Camelid somatic affinity variants yields ultra-potent human therapeutic IL-6 antibodies
To be Published
|
|
3QI8
 
 | | Evolved variant of cytochrome P450 (BM3, CYP102A1) | | Descriptor: | Evolved Cytochrome P450 variant (22A3), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Rentmeister, A, Brown, T.R, Snow, C.D, Carbone, M.N, Arnold, F.H. | | Deposit date: | 2011-01-26 | | Release date: | 2011-05-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Engineered Bacterial Mimics of Human Drug Metabolizing Enzyme CYP2C9
Chemcatchem, 2011
|
|
3DPA
 
 | |
2ESG
 
 | | Solution structure of the complex between immunoglobulin IgA1 and human serum albumin | | Descriptor: | Immunoglobulin A1 heavy chain, Immunoglobulin A1 light chain, Serum albumin | | Authors: | Almogren, A, Furtado, P.B, Sun, Z, Perkins, S.J, Kerr, M.A. | | Deposit date: | 2005-10-26 | | Release date: | 2006-01-31 | | Last modified: | 2024-02-14 | | Method: | SOLUTION SCATTERING | | Cite: | Purification, Properties and Extended Solution Structure of the Complex Formed between Human Immunoglobulin A1 and Human Serum Albumin by Scattering and Ultracentrifugation.
J.Mol.Biol., 356, 2006
|
|
1QL3
 
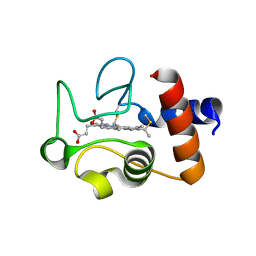 | | Structure of the soluble domain of cytochrome c552 from Paracoccus denitrificans in the reduced state | | Descriptor: | CYTOCHROME C552, HEME C | | Authors: | Harrenga, A, Reincke, B, Rueterjans, H, Ludwig, B, Michel, H. | | Deposit date: | 1999-08-20 | | Release date: | 2000-02-06 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of the Soluble Domain of Cytochrome C552 from Paracoccus Denitrificans in the Oxidized and Reduced States
J.Mol.Biol., 295, 2000
|
|
1QL4
 
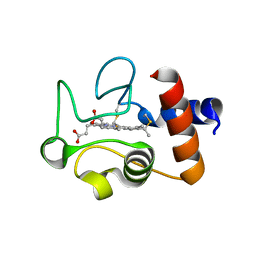 | | Structure of the soluble domain of cytochrome c552 from Paracoccus denitrificans in the oxidised state | | Descriptor: | CYTOCHROME C552, HEME C | | Authors: | Harrenga, A, Reincke, B, Rueterjans, H, Ludwig, B, Michel, H. | | Deposit date: | 1999-08-20 | | Release date: | 2000-02-03 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of the Soluble Domain of Cytochrome C552 from Paracoccus Denitrificans in the Oxidized and Reduced States
J.Mol.Biol., 295, 2000
|
|
1QLE
 
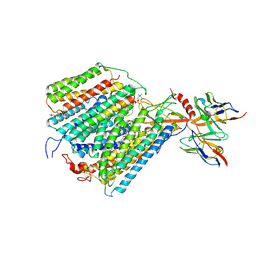 | |
