4M92
 
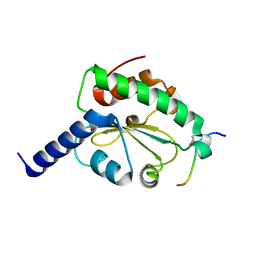 | | Crystal structure of hN33/Tusc3-peptide 2 | | Descriptor: | Interleukin-1 receptor accessory protein-like 1, Tumor suppressor candidate 3 | | Authors: | Mohorko, E, Owen, R.L, Malojcic, G, Brozzo, M.S, Aebi, M, Glockshuber, R. | | Deposit date: | 2013-08-14 | | Release date: | 2014-03-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis of substrate specificity of human oligosaccharyl transferase subunit n33/tusc3 and its role in regulating protein N-glycosylation.
Structure, 22, 2014
|
|
4M91
 
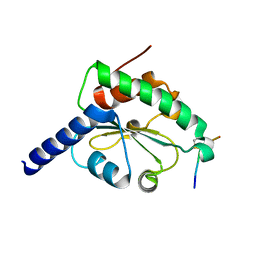 | | crystal structure of hN33/Tusc3-peptide 1 | | Descriptor: | Protein cereblon, Tumor suppressor candidate 3 | | Authors: | Mohorko, E, Owen, R.L, Malojcic, G, Brozzo, M.S, Aebi, M, Glockshuber, R. | | Deposit date: | 2013-08-14 | | Release date: | 2014-03-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structural basis of substrate specificity of human oligosaccharyl transferase subunit n33/tusc3 and its role in regulating protein N-glycosylation.
Structure, 22, 2014
|
|
1HFP
 
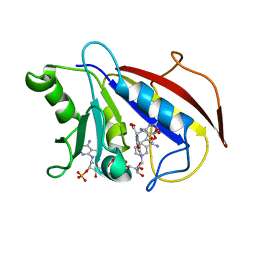 | | COMPARISON OF TERNARY CRYSTAL COMPLEXES OF HUMAN DIHYDROFOLATE REDUCTASE WITH NADPH AND A CLASSICAL ANTITUMOR FUROPYRIMDINE | | Descriptor: | DIHYDROFOLATE REDUCTASE, N-[4-[(2,4-DIAMINOFURO[2,3D]PYRIMIDIN-5-YL)METHYL]METHYLAMINO]-BENZOYL]-L-GLUTAMATE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cody, V, Galitsky, N, Luft, J.R, Pangborn, W, Blakley, R.L, Gangjee, A. | | Deposit date: | 1997-11-04 | | Release date: | 1998-01-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Comparison of ternary crystal complexes of F31 variants of human dihydrofolate reductase with NADPH and a classical antitumor furopyrimidine.
Anti-Cancer Drug Des., 13, 1998
|
|
1HFQ
 
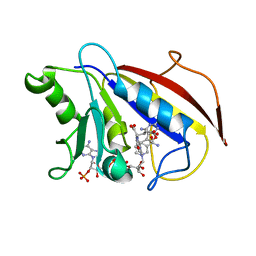 | | COMPARISON OF TERNARY CRYSTAL COMPLEXES OF HUMAN DIHYDROFOLATE REDUCTASE WITH NADPH AND A CLASSICAL ANTITUMOR FUROPYRIMDINE | | Descriptor: | DIHYDROFOLATE REDUCTASE, N-[4-[(2,4-DIAMINOFURO[2,3D]PYRIMIDIN-5-YL)METHYL]METHYLAMINO]-BENZOYL]-L-GLUTAMATE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cody, V, Galitsky, N, Luft, J.R, Pangborn, W, Blakley, R.L, Gangjee, A. | | Deposit date: | 1997-11-04 | | Release date: | 1998-01-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Comparison of ternary crystal complexes of F31 variants of human dihydrofolate reductase with NADPH and a classical antitumor furopyrimidine.
Anti-Cancer Drug Des., 13, 1998
|
|
1HFR
 
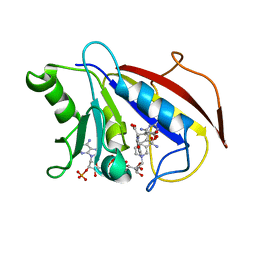 | | COMPARISON OF TERNARY CRYSTAL COMPLEXES OF HUMAN DIHYDROFOLATE REDUCTASE WITH NADPH AND A CLASSICAL ANTITUMOR FUROPYRIMDINE | | Descriptor: | DIHYDROFOLATE REDUCTASE, N-[4-[(2,4-DIAMINOFURO[2,3D]PYRIMIDIN-5-YL)METHYL]METHYLAMINO]-BENZOYL]-L-GLUTAMATE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cody, V, Galitsky, N, Luft, J.R, Pangborn, W, Blakley, R.L, Gangjee, A. | | Deposit date: | 1997-11-04 | | Release date: | 1998-01-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Comparison of ternary crystal complexes of F31 variants of human dihydrofolate reductase with NADPH and a classical antitumor furopyrimidine.
Anti-Cancer Drug Des., 13, 1998
|
|
6VIO
 
 | |
6E3C
 
 | |
6EYO
 
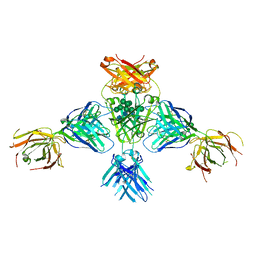 | | Structure of extended IgE-Fc in complex with two anti-IgE Fabs | | Descriptor: | 8D6 Fab heavy chain, 8D6 Fab light chain, Immunoglobulin heavy constant epsilon, ... | | Authors: | Chen, J.B, Ramadani, F, Pang, M.O.Y, Beavil, R.L, Holdom, M.D, Mitropoulou, A.N, Beavil, A.J, Gould, H.J, Chang, T.W, Sutton, B.J, McDonnell, J.M, Davies, A.M. | | Deposit date: | 2017-11-13 | | Release date: | 2018-08-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structural basis for selective inhibition of immunoglobulin E-receptor interactions by an anti-IgE antibody.
Sci Rep, 8, 2018
|
|
6EYN
 
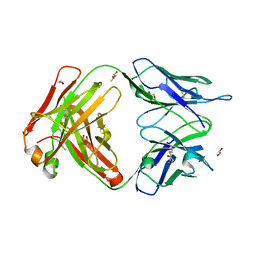 | | Structure of the 8D6 (anti-IgE) Fab | | Descriptor: | 1,2-ETHANEDIOL, 8D6 Fab heavy chain, 8D6 Fab light chain, ... | | Authors: | Chen, J.B, Ramadani, F, Pang, M.O.Y, Beavil, R.L, Holdom, M.D, Mitropoulou, A.N, Beavil, A.J, Gould, H.J, Chang, T.W, Sutton, B.J, McDonnell, J.M, Davies, A.M. | | Deposit date: | 2017-11-13 | | Release date: | 2018-08-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for selective inhibition of immunoglobulin E-receptor interactions by an anti-IgE antibody.
Sci Rep, 8, 2018
|
|
2AJL
 
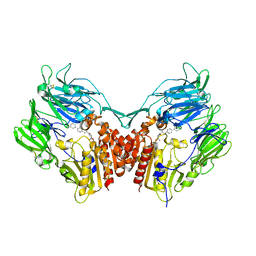 | | X-ray Structure of Novel Biaryl-Based Dipeptidyl peptidase IV inhibitor | | Descriptor: | 1-[2-(S)-AMINO-3-BIPHENYL-4-YL-PROPIONYL]-PYRROLIDINE-2-(S)-CARBONITRILE, 2-acetamido-2-deoxy-beta-D-glucopyranose, Dipeptidyl peptidase 4 | | Authors: | Qiao, L, Baumann, C.A, Crysler, C.S, Ninan, N.S, Abad, M.C, Spurlino, J.C, DesJarlais, R.L, Kervinen, J, Neeper, M.P, Bayoumy, S.S. | | Deposit date: | 2005-08-02 | | Release date: | 2005-11-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery, SAR, and X-ray structure of novel biaryl-based dipeptidyl peptidase IV inhibitors
Bioorg.Med.Chem.Lett., 16, 2006
|
|
8B3S
 
 | | Structure of YjbA in complex with ClpC N-terminal Domain | | Descriptor: | ATP-dependent Clp protease ATP-binding subunit ClpC / Negative regulator of tic competence clcC/mecB, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Evans, N.J, Isaacson, R.L, Camp, A.H, Collins, M.K. | | Deposit date: | 2022-09-16 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structure of YjbA in complex with ClpC N-terminal Domain
To Be Published
|
|
5FCX
 
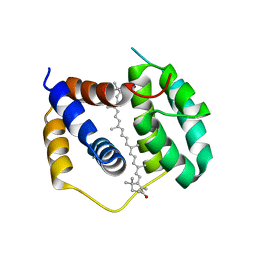 | | Structure of Anabaena (Nostoc) sp. PCC 7120 Red Carotenoid Protein binding canthaxanthin | | Descriptor: | Red carotenoid protein (RCP), beta,beta-carotene-4,4'-dione | | Authors: | Sutter, M, Leverenz, R.L, Kerfeld, C.A. | | Deposit date: | 2015-12-15 | | Release date: | 2016-07-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.206 Å) | | Cite: | Structure, Diversity, and Evolution of a New Family of Soluble Carotenoid-Binding Proteins in Cyanobacteria.
Mol Plant, 9, 2016
|
|
5FGT
 
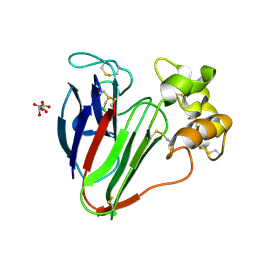 | | Thaumatin solved by native sulphur-SAD using free-electron laser radiation | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin-1 | | Authors: | Nass, K.J, Meinhart, A, Barends, T.R.M, Foucar, L, Gorel, A, Aquila, A, Botha, S, Doak, R.B, Koglin, J, Liang, M, Shoeman, R.L, Williams, G.K, Boutet, S, Schlichting, I. | | Deposit date: | 2015-12-21 | | Release date: | 2016-06-08 | | Last modified: | 2018-11-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Protein structure determination by single-wavelength anomalous diffraction phasing of X-ray free-electron laser data.
Iucrj, 3, 2016
|
|
5FGX
 
 | | Thaumatin solved by native sulphur SAD using synchrotron radiation | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin-1 | | Authors: | Nass, K.J, Meinhart, A, Barends, T.R.M, Foucar, L, Gorel, A, Aquila, A, Botha, S, Doak, R.B, Koglin, J, Liang, M, Shoeman, R.L, Williams, G.J, Boutet, S, Schlichting, I. | | Deposit date: | 2015-12-21 | | Release date: | 2016-06-08 | | Method: | X-RAY DIFFRACTION (2.134 Å) | | Cite: | Protein structure determination by single-wavelength anomalous diffraction phasing of X-ray free-electron laser data.
Iucrj, 3, 2016
|
|
7RTX
 
 | | Actophorin grown in microgravity | | Descriptor: | Actophorin | | Authors: | Lieberman, R.L, Quirk, S. | | Deposit date: | 2021-08-16 | | Release date: | 2021-12-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Improved resolution crystal structure of Acanthamoeba actophorin reveals structural plasticity not induced by microgravity.
Acta Crystallogr.,Sect.F, 77, 2021
|
|
7RTG
 
 | | Crystal Structure of the Human Adenosine Deaminase 1 | | Descriptor: | Adenosine deaminase, ZINC ION | | Authors: | Ma, M.T, Lieberman, R.L, Blazeck, J, Jennings, M.R. | | Deposit date: | 2021-08-13 | | Release date: | 2022-01-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.591 Å) | | Cite: | Catalytically active holo Homo sapiens adenosine deaminase I adopts a closed conformation.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
4W8A
 
 | | Crystal structure of XEG5B, a GH5 xyloglucan-specific beta-1,4-glucanase from ruminal metagenomic library, in the native form | | Descriptor: | Exo-xyloglucanase, GLYCEROL, SULFATE ION | | Authors: | Santos, C.R, Cordeiro, R.L, Wong, D.W.S, Murakami, M.T. | | Deposit date: | 2014-08-22 | | Release date: | 2015-03-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structural Basis for Xyloglucan Specificity and alpha-d-Xylp(1 6)-d-Glcp Recognition at the -1 Subsite within the GH5 Family.
Biochemistry, 54, 2015
|
|
4W85
 
 | | Crystal structure of XEG5A, a GH5 xyloglucan-specific endo-beta-1,4-glucanase from ruminal metagenomic library, in complex with glucose | | Descriptor: | MAGNESIUM ION, Xyloglucan-specific endo-beta-1,4-glucanase, beta-D-glucopyranose | | Authors: | Santos, C.R, Cordeiro, R.L, Wong, D.W.S, Murakami, M.T. | | Deposit date: | 2014-08-22 | | Release date: | 2015-03-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural Basis for Xyloglucan Specificity and alpha-d-Xylp(1 6)-d-Glcp Recognition at the -1 Subsite within the GH5 Family.
Biochemistry, 54, 2015
|
|
4W84
 
 | | Crystal structure of XEG5A, a GH5 xyloglucan-specific endo-beta-1,4-glucanase from ruminal metagenomic library, in the native form | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MAGNESIUM ION, Xyloglucan-specific endo-beta-1,4-glucanase | | Authors: | Santos, C.R, Cordeiro, R.L, Wong, D.W.S, Murakami, M.T. | | Deposit date: | 2014-08-22 | | Release date: | 2015-03-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural Basis for Xyloglucan Specificity and alpha-d-Xylp(1 6)-d-Glcp Recognition at the -1 Subsite within the GH5 Family.
Biochemistry, 54, 2015
|
|
4W87
 
 | | Crystal structure of XEG5A, a GH5 xyloglucan-specific endo-beta-1,4-glucanase from metagenomic library, in complex with a xyloglucan oligosaccharide | | Descriptor: | MAGNESIUM ION, Xyloglucan-specific endo-beta-1,4-glucanase, alpha-D-xylopyranose-(1-6)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Santos, C.R, Cordeiro, R.L, Wong, D.W.S, Murakami, M.T. | | Deposit date: | 2014-08-22 | | Release date: | 2015-03-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural Basis for Xyloglucan Specificity and alpha-d-Xylp(1 6)-d-Glcp Recognition at the -1 Subsite within the GH5 Family.
Biochemistry, 54, 2015
|
|
1QK5
 
 | | TOXOPLASMA GONDII HYPOXANTHINE-GUANINE PHOSPHORIBOSYLTRANSFERASE WITH XMP, PYROPHOSPHATE AND TWO MG2+ IONS | | Descriptor: | HYPOXANTHINE-GUANINE PHOSPHORIBOSYLTRANSFERASE, MAGNESIUM ION, PYROPHOSPHATE 2-, ... | | Authors: | Heroux, A, White, E.L, Ross, L.J, Davis, R.L, Borhani, D.W. | | Deposit date: | 1999-07-09 | | Release date: | 1999-10-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of Toxoplasma Gondii Hypoxanthine-Guanine Phosphoribosyltransferase with Xmp, Pyrophosphate and Two Mg2+ Ions Bound: Insights Into the Catalytic Mechanism
Biochemistry, 38, 1999
|
|
1HGH
 
 | | BINDING OF INFLUENZA VIRUS HEMAGGLUTININ TO ANALOGS OF ITS CELL-SURFACE RECEPTOR, SIALIC ACID: ANALYSIS BY PROTON NUCLEAR MAGNETIC RESONANCE SPECTROSCOPY AND X-RAY CRYSTALLOGRAPHY | | Descriptor: | 2-O-methyl-5-N-acetyl-alpha-D-neuraminic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, HEMAGGLUTININ, ... | | Authors: | Sauter, N.K, Hanson, J.E, Glick, G.D, Brown, J.H, Crowther, R.L, Park, S.-J, Skehel, J.J, Wiley, D.C. | | Deposit date: | 1991-11-01 | | Release date: | 1994-01-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Binding of influenza virus hemagglutinin to analogs of its cell-surface receptor, sialic acid: analysis by proton nuclear magnetic resonance spectroscopy and X-ray crystallography.
Biochemistry, 31, 1992
|
|
1HGG
 
 | | BINDING OF INFLUENZA VIRUS HEMAGGLUTININ TO ANALOGS OF ITS CELL-SURFACE RECEPTOR, SIALIC ACID: ANALYSIS BY PROTON NUCLEAR MAGNETIC RESONANCE SPECTROSCOPY AND X-RAY CRYSTALLOGRAPHY | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HEMAGGLUTININ, CHAIN HA1, ... | | Authors: | Sauter, N.K, Hanson, J.E, Glick, G.D, Brown, J.H, Crowther, R.L, Park, S.-J, Skehel, J.J, Wiley, D.C. | | Deposit date: | 1991-11-01 | | Release date: | 1994-01-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Binding of influenza virus hemagglutinin to analogs of its cell-surface receptor, sialic acid: analysis by proton nuclear magnetic resonance spectroscopy and X-ray crystallography.
Biochemistry, 31, 1992
|
|
1HGJ
 
 | | BINDING OF INFLUENZA VIRUS HEMAGGLUTININ TO ANALOGS OF ITS CELL-SURFACE RECEPTOR, SIALIC ACID: ANALYSIS BY PROTON NUCLEAR MAGNETIC RESONANCE SPECTROSCOPY AND X-RAY CRYSTALLOGRAPHY | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HEMAGGLUTININ, CHAIN HA1, ... | | Authors: | Sauter, N.K, Hanson, J.E, Glick, G.D, Brown, J.H, Crowther, R.L, Park, S.-J, Skehel, J.J, Wiley, D.C. | | Deposit date: | 1991-11-01 | | Release date: | 1994-01-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Binding of influenza virus hemagglutinin to analogs of its cell-surface receptor, sialic acid: analysis by proton nuclear magnetic resonance spectroscopy and X-ray crystallography.
Biochemistry, 31, 1992
|
|
3KBL
 
 | | Crystal structure of the GLD-1 homodimerization domain from Caenorhabditis elegans N169A mutant at 2.28 A resolution | | Descriptor: | Female germline-specific tumor suppressor gld-1 | | Authors: | Beuck, C, Szymczyna, B.R, Kerkow, D.E, Carmel, A.B, Columbus, L, Stanfield, R.L, Williamson, J.R. | | Deposit date: | 2009-10-20 | | Release date: | 2010-03-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structure of the GLD-1 homodimerization domain: insights into STAR protein-mediated translational regulation.
Structure, 18, 2010
|
|
