8HBL
 
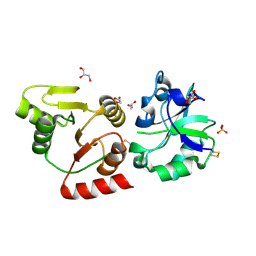 | | Crystal structure of the SARS-unique domain (SUD) of SARS-CoV-2 (1.58 angstrom resolution) | | Descriptor: | GLYCEROL, LITHIUM ION, Non-structural protein 3, ... | | Authors: | Qin, B, Li, Z, Aumonier, S, Wang, M, Cui, S. | | Deposit date: | 2022-10-29 | | Release date: | 2023-07-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Identification of the SARS-unique domain of SARS-CoV-2 as an antiviral target.
Nat Commun, 14, 2023
|
|
8GQC
 
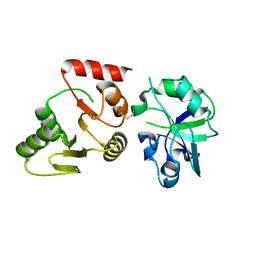 | | Crystal structure of the SARS-unique domain (SUD) of SARS-CoV-2 (1.35 angstrom resolution) | | Descriptor: | Papain-like protease nsp3 | | Authors: | Qin, B, Li, Z, Aumonier, S, Wang, M, Cui, S. | | Deposit date: | 2022-08-30 | | Release date: | 2023-07-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Identification of the SARS-unique domain of SARS-CoV-2 as an antiviral target.
Nat Commun, 14, 2023
|
|
7WYO
 
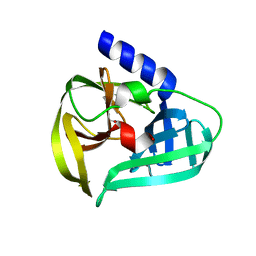 | | Structure of the EV71 3Cpro with 338 inhibitor | | Descriptor: | 3C protein, N-methyl-N-(4,5,6,7-tetrahydro-1,3-benzothiazol-2-ylmethyl)prop-2-enamide | | Authors: | Qin, B, Hou, P, Gao, X, Cui, S. | | Deposit date: | 2022-02-16 | | Release date: | 2022-06-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.402 Å) | | Cite: | Acrylamide fragment inhibitors that induce unprecedented conformational distortions in enterovirus 71 3C and SARS-CoV-2 main protease.
Acta Pharm Sin B, 12, 2022
|
|
7WYM
 
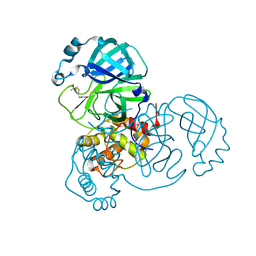 | | Structure of the SARS-COV-2 main protease with 337 inhibitor | | Descriptor: | 3C-like proteinase nsp5, N-methyl-N-[[4-(trifluoromethyl)-1,3-thiazol-2-yl]methyl]prop-2-enamide | | Authors: | Qin, B, Hou, P, Gao, X, Cui, S. | | Deposit date: | 2022-02-16 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Acrylamide fragment inhibitors that induce unprecedented conformational distortions in enterovirus 71 3C and SARS-CoV-2 main protease.
Acta Pharm Sin B, 12, 2022
|
|
7WYL
 
 | | Structure of the EV71 3Cpro with 337 inhibitor | | Descriptor: | 3C protein, N-methyl-N-[[4-(trifluoromethyl)-1,3-thiazol-2-yl]methyl]prop-2-enamide | | Authors: | Qin, B, Hou, P, Gao, X, Cui, S. | | Deposit date: | 2022-02-16 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Acrylamide fragment inhibitors that induce unprecedented conformational distortions in enterovirus 71 3C and SARS-CoV-2 main protease.
Acta Pharm Sin B, 12, 2022
|
|
7WYP
 
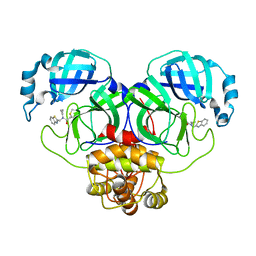 | | Structure of the SARS-COV-2 main protease with EN102 inhibitor | | Descriptor: | 3C-like proteinase, N-(1,3-benzothiazol-2-ylmethyl)-N-cyclopropyl-prop-2-enamide | | Authors: | Qin, B, Hou, P, Gao, X, Cui, S. | | Deposit date: | 2022-02-16 | | Release date: | 2022-06-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Acrylamide fragment inhibitors that induce unprecedented conformational distortions in enterovirus 71 3C and SARS-CoV-2 main protease.
Acta Pharm Sin B, 12, 2022
|
|
5B6K
 
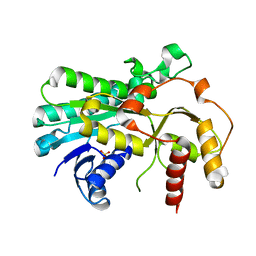 | |
8C8Y
 
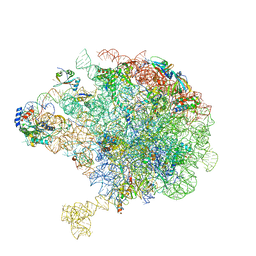 | | Cryo-EM captures early ribosome assembly in action | | Descriptor: | 23S rRNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Lauer, S, Nikolay, R, Qin, B. | | Deposit date: | 2023-01-21 | | Release date: | 2023-04-05 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Cryo-EM captures early ribosome assembly in action.
Nat Commun, 14, 2023
|
|
8C94
 
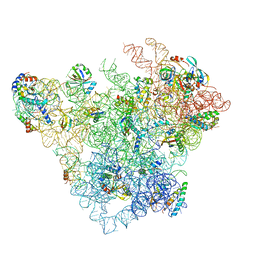 | | Cryo-EM captures early ribosome assembly in action | | Descriptor: | 23S rRNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Lauer, S, Nikolay, R, Qin, B. | | Deposit date: | 2023-01-21 | | Release date: | 2023-04-05 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM captures early ribosome assembly in action.
Nat Commun, 14, 2023
|
|
8C98
 
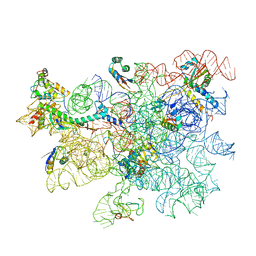 | | Cryo-EM captures early ribosome assembly in action | | Descriptor: | 23S rRNA, 50S ribosomal protein L13, 50S ribosomal protein L15, ... | | Authors: | Lauer, S, Nikolay, R, Qin, B. | | Deposit date: | 2023-01-21 | | Release date: | 2023-04-05 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | Cryo-EM captures early ribosome assembly in action.
Nat Commun, 14, 2023
|
|
8C97
 
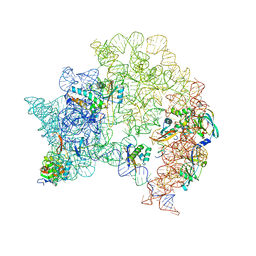 | | Cryo-EM captures early ribosome assembly in action | | Descriptor: | 23S rRNA, 50S ribosomal protein L14, 50S ribosomal protein L17, ... | | Authors: | Lauer, S, Nikolay, R, Qin, B. | | Deposit date: | 2023-01-21 | | Release date: | 2023-04-05 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.07 Å) | | Cite: | Cryo-EM captures early ribosome assembly in action.
Nat Commun, 14, 2023
|
|
8C90
 
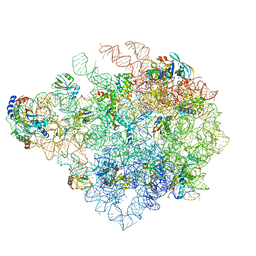 | | Cryo-EM captures early ribosome assembly in action | | Descriptor: | 23S rRNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Lauer, S, Nikolay, R, Qin, B. | | Deposit date: | 2023-01-21 | | Release date: | 2023-04-05 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Cryo-EM captures early ribosome assembly in action.
Nat Commun, 14, 2023
|
|
8C9A
 
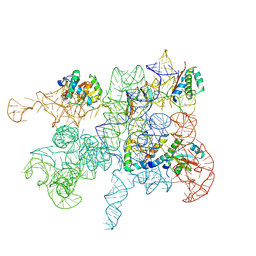 | | Cryo-EM captures early ribosome assembly in action | | Descriptor: | 23S rRNA, 50S ribosomal protein L22, 50S ribosomal protein L23, ... | | Authors: | Lauer, S, Nikolay, R, Qin, B. | | Deposit date: | 2023-01-21 | | Release date: | 2023-04-05 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.86 Å) | | Cite: | Cryo-EM captures early ribosome assembly in action.
Nat Commun, 14, 2023
|
|
8C8X
 
 | | Cryo-EM captures early ribosome assembly in action | | Descriptor: | 23S rRNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Lauer, S, Nikolay, R, Qin, B. | | Deposit date: | 2023-01-21 | | Release date: | 2023-04-05 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.93 Å) | | Cite: | Cryo-EM captures early ribosome assembly in action.
Nat Commun, 14, 2023
|
|
8C95
 
 | | Cryo-EM captures early ribosome assembly in action | | Descriptor: | 23S rRNA, 50S ribosomal protein L13, 50S ribosomal protein L15, ... | | Authors: | Lauer, S, Nikolay, R, Qin, B. | | Deposit date: | 2023-01-21 | | Release date: | 2023-04-05 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.92 Å) | | Cite: | Cryo-EM captures early ribosome assembly in action.
Nat Commun, 14, 2023
|
|
8C96
 
 | | Cryo-EM captures early ribosome assembly in action | | Descriptor: | 23S rRNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Lauer, S, Nikolay, R, Qin, B. | | Deposit date: | 2023-01-21 | | Release date: | 2023-04-05 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.43 Å) | | Cite: | Cryo-EM captures early ribosome assembly in action.
Nat Commun, 14, 2023
|
|
8C9B
 
 | | Cryo-EM captures early ribosome assembly in action | | Descriptor: | 23S rRNA, 50S ribosomal protein L22, 50S ribosomal protein L24, ... | | Authors: | Nikolay, R, Qin, B, Lauer, S. | | Deposit date: | 2023-01-21 | | Release date: | 2023-04-05 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | Cryo-EM captures early ribosome assembly in action.
Nat Commun, 14, 2023
|
|
8C93
 
 | | Cryo-EM captures early ribosome assembly in action | | Descriptor: | 23S rRNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Lauer, S, Nikolay, R, Qin, B. | | Deposit date: | 2023-01-21 | | Release date: | 2023-04-05 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.17 Å) | | Cite: | Cryo-EM captures early ribosome assembly in action.
Nat Commun, 14, 2023
|
|
8C99
 
 | | Cryo-EM captures early ribosome assembly in action | | Descriptor: | 23S rRNA, 50S ribosomal protein L13, 50S ribosomal protein L15, ... | | Authors: | Nikolay, R, Qin, B, Lauer, S. | | Deposit date: | 2023-01-21 | | Release date: | 2023-04-05 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Cryo-EM captures early ribosome assembly in action.
Nat Commun, 14, 2023
|
|
8C9C
 
 | | Cryo-EM captures early ribosome assembly in action | | Descriptor: | 23S rRNA, 50S ribosomal protein L22, 50S ribosomal protein L24, ... | | Authors: | Nikolay, R, Qin, B, Lauer, S. | | Deposit date: | 2023-01-21 | | Release date: | 2023-04-05 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (6.62 Å) | | Cite: | Cryo-EM captures early ribosome assembly in action.
Nat Commun, 14, 2023
|
|
8C91
 
 | | Cryo-EM captures early ribosome assembly in action | | Descriptor: | 23S rRNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Lauer, S, Nikolay, R, Qin, B. | | Deposit date: | 2023-01-21 | | Release date: | 2023-04-05 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.19 Å) | | Cite: | Cryo-EM captures early ribosome assembly in action.
Nat Commun, 14, 2023
|
|
8C8Z
 
 | | Cryo-EM captures early ribosome assembly in action | | Descriptor: | 23S rRNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Lauer, S, Nikolay, R, Qin, B. | | Deposit date: | 2023-01-21 | | Release date: | 2023-04-05 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Cryo-EM captures early ribosome assembly in action.
Nat Commun, 14, 2023
|
|
8C92
 
 | | Cryo-EM captures early ribosome assembly in action | | Descriptor: | 23S rRNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Lauer, S, Nikolay, R, Qin, B. | | Deposit date: | 2023-01-21 | | Release date: | 2023-04-05 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.79 Å) | | Cite: | Cryo-EM captures early ribosome assembly in action.
Nat Commun, 14, 2023
|
|
6GBZ
 
 | | 50S ribosomal subunit assembly intermediate state 5 | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Nikolay, R, Hilal, T, Qin, B, Loerke, J, Buerger, J, Mielke, T, Spahn, C.M.T. | | Deposit date: | 2018-04-16 | | Release date: | 2018-06-20 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural Visualization of the Formation and Activation of the 50S Ribosomal Subunit during In Vitro Reconstitution.
Mol. Cell, 70, 2018
|
|
6GC8
 
 | | 50S ribosomal subunit assembly intermediate - 50S rec* | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Nikolay, R, Hilal, T, Qin, B, Loerke, J, Buerger, J, Mielke, T, Spahn, C.M.T. | | Deposit date: | 2018-04-17 | | Release date: | 2018-06-20 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural Visualization of the Formation and Activation of the 50S Ribosomal Subunit during In Vitro Reconstitution.
Mol. Cell, 70, 2018
|
|
