6E8R
 
 | |
7SR1
 
 | |
7SR2
 
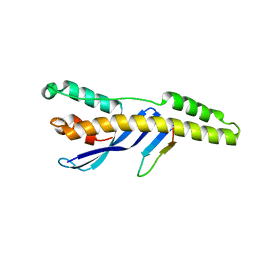
 | | Crystal structure of the human SNX25 regulator of G-protein signalling (RGS) domain | | Descriptor: | ACETATE ION, LEUCINE, Sorting nexin-25, ... | | Authors: | Collins, B.M, Paul, B, Weeratunga, S. | | Deposit date: | 2021-11-07 | | Release date: | 2021-11-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structural Predictions of the SNX-RGS Proteins Suggest They Belong to a New Class of Lipid Transfer Proteins.
Front Cell Dev Biol, 10, 2022
|
|
5TGH
 
 | |
3HKN
 
 | | Human carbonic anhydrase II in complex with (2,3,4,6-Tetra-O-acetyl-beta-D-galactopyranosyl) -(1-4)-1,2,3,6-tetra-O-acetyl-1-thio-beta-D-glucopyranosylsulfonamide | | Descriptor: | 2,3,4,6-tetra-O-acetyl-beta-D-galactopyranose-(1-4)-(1S)-2,3,6-tri-O-acetyl-1,5-anhydro-1-sulfamoyl-D-glucitol, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Paul, B, Poulsen, S.-A, Hofmann, A. | | Deposit date: | 2009-05-25 | | Release date: | 2009-10-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | S-glycosyl primary sulfonamides--a new structural class for selective inhibition of cancer-associated carbonic anhydrases.
J.Med.Chem., 52, 2009
|
|
3HKQ
 
 | | Human carbonic anhydrase II in complex with 1-S-D-Galactopyranosylsulfonamide | | Descriptor: | (1S)-1,5-anhydro-1-sulfamoyl-D-galactitol, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Paul, B, Poulsen, S.-A, Hofmann, A. | | Deposit date: | 2009-05-25 | | Release date: | 2009-10-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | S-glycosyl primary sulfonamides--a new structural class for selective inhibition of cancer-associated carbonic anhydrases.
J.Med.Chem., 52, 2009
|
|
3HKU
 
 | | Human carbonic anhydrase II in complex with topiramate | | Descriptor: | Carbonic anhydrase 2, ZINC ION, [(3aS,5aR,8aR,8bS)-2,2,7,7-tetramethyltetrahydro-3aH-bis[1,3]dioxolo[4,5-b:4',5'-d]pyran-3a-yl]methyl sulfamate | | Authors: | Paul, B, Poulsen, S.-A, Hofmann, A. | | Deposit date: | 2009-05-25 | | Release date: | 2009-10-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | S-glycosyl primary sulfonamides--a new structural class for selective inhibition of cancer-associated carbonic anhydrases.
J.Med.Chem., 52, 2009
|
|
3HKT
 
 | | Human carbonic anhydrase II in complex with alpha-D-Glucopyranosyl-(1->4)-1-thio-beta-D-glucopyranosylsulfonamide | | Descriptor: | Carbonic anhydrase 2, ZINC ION, alpha-D-galactopyranose-(1-4)-(1S)-1,5-anhydro-1-sulfamoyl-D-galactitol | | Authors: | Paul, B, Poulsen, S.-A, Hofmann, A. | | Deposit date: | 2009-05-25 | | Release date: | 2009-10-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | S-glycosyl primary sulfonamides--a new structural class for selective inhibition of cancer-associated carbonic anhydrases.
J.Med.Chem., 52, 2009
|
|
6N5X
 
 | | Crystal structure of the SNX5 PX domain in complex with the CI-MPR (space group P212121 - Form 1) | | Descriptor: | GLYCEROL, O-(O-(2-AMINOPROPYL)-O'-(2-METHOXYETHYL)POLYPROPYLENE GLYCOL 500), Sorting nexin-5,Cation-independent mannose-6-phosphate receptor | | Authors: | Collins, B, Paul, B, Weeratunga, S. | | Deposit date: | 2018-11-22 | | Release date: | 2019-09-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.051 Å) | | Cite: | Molecular identification of a BAR domain-containing coat complex for endosomal recycling of transmembrane proteins.
Nat.Cell Biol., 21, 2019
|
|
6N5Z
 
 | |
6N5Y
 
 | |
7CZC
 
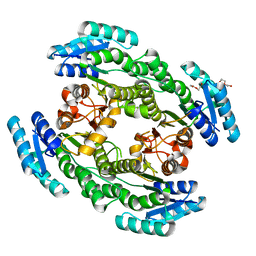 | | Crystal structure of apo-FabG from Vibrio harveyi | | Descriptor: | 3-oxoacyl-ACP reductase FabG, DI(HYDROXYETHYL)ETHER | | Authors: | Singh, B.K, Kumar, A, Paul, B, Biswas, R, Das, A.K. | | Deposit date: | 2020-09-08 | | Release date: | 2021-09-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of apo-FabG from Vibrio harveyi
To Be Published
|
|
5TGJ
 
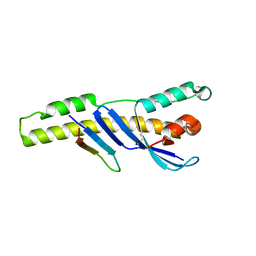 | |
5TGI
 
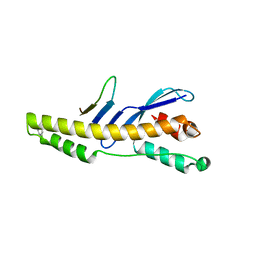 | |
4TW3
 
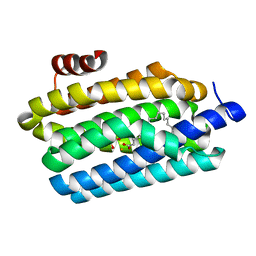 | | Insights into Substrate and Metal Binding from the Crystal Structure of Cyanobacterial Aldehyde Deformylating Oxygenase with Substrate Bound | | Descriptor: | 1,2-ETHANEDIOL, Aldehyde decarbonylase, FE (III) ION, ... | | Authors: | Buer, B.C, Paul, B, Das, D, Stuckey, J.A, Marsh, E.N.G. | | Deposit date: | 2014-06-29 | | Release date: | 2014-10-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Insights into substrate and metal binding from the crystal structure of cyanobacterial aldehyde deformylating oxygenase with substrate bound.
Acs Chem.Biol., 9, 2014
|
|
6ERI
 
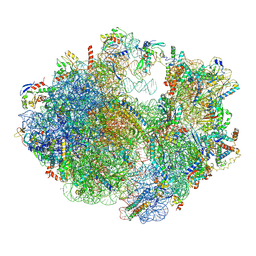 | | Structure of the chloroplast ribosome with chl-RRF and hibernation-promoting factor | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10 alpha, ... | | Authors: | Perez Borema, A, Aibara, S, Paul, B, Tobiasson, V, Kimanius, D, Forsberg, B.O, Wallden, K, Lindahl, E, Amunts, A. | | Deposit date: | 2017-10-18 | | Release date: | 2018-04-18 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of the chloroplast ribosome with chl-RRF and hibernation-promoting factor.
Nat Plants, 4, 2018
|
|
4PGK
 
 | | Insights into Substrate and Metal Binding from the Crystal Structure of Cyanobacterial Aldehyde Deformylating Oxygenase with Substrate Bound | | Descriptor: | 11-[2-(2-ethoxyethoxy)ethoxy]undecanal, Aldehyde decarbonylase, FE (III) ION | | Authors: | Buer, B.C, Paul, B, Das, D, Stuckey, J.A, Marsh, E.N.G. | | Deposit date: | 2014-05-02 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Insights into substrate and metal binding from the crystal structure of cyanobacterial aldehyde deformylating oxygenase with substrate bound.
Acs Chem.Biol., 9, 2014
|
|
4PGI
 
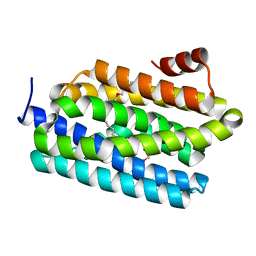 | | Insights into Substrate and Metal Binding from the Crystal Structure of Cyanobacterial Aldehyde Deformylating Oxygenase with Substrate Analogs Bound | | Descriptor: | 1,2-ETHANEDIOL, 11-[2-(2-ethoxyethoxy)ethoxy]undecanal, Aldehyde decarbonylase, ... | | Authors: | Buer, B.C, Paul, B, Das, D, Stuckey, J.A, Marsh, E.N.G. | | Deposit date: | 2014-05-02 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Insights into substrate and metal binding from the crystal structure of cyanobacterial aldehyde deformylating oxygenase with substrate bound.
Acs Chem.Biol., 9, 2014
|
|
4PG1
 
 | | Insights into Substrate and Metal Binding from the Crystal Structure of Cyanobacterial Aldehyde Deformylating Oxygenase with Substrate Bound | | Descriptor: | (1S,2S)-2-nonylcyclopropanecarboxylic acid, Aldehyde decarbonylase, DIMETHYL SULFOXIDE, ... | | Authors: | Buer, B.C, Paul, B, Das, D, Stuckey, J.A, Marsh, E.N.G. | | Deposit date: | 2014-05-01 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Insights into substrate and metal binding from the crystal structure of cyanobacterial aldehyde deformylating oxygenase with substrate bound.
Acs Chem.Biol., 9, 2014
|
|
4PG0
 
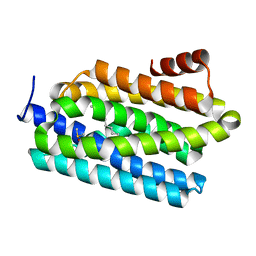 | | Insights into Substrate and Metal Binding from the Crystal Structure of Cyanobacterial Aldehyde Deformylating Oxygenase with Substrate Bound | | Descriptor: | (1S,2S)-2-nonylcyclopropanecarboxylic acid, Aldehyde decarbonylase, DIMETHYL SULFOXIDE, ... | | Authors: | Buer, B.C, Paul, B, Das, D, Stuckey, J.A, Marsh, E.N.G. | | Deposit date: | 2014-05-01 | | Release date: | 2014-11-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Insights into substrate and metal binding from the crystal structure of cyanobacterial aldehyde deformylating oxygenase with substrate bound.
Acs Chem.Biol., 9, 2014
|
|
5IOO
 
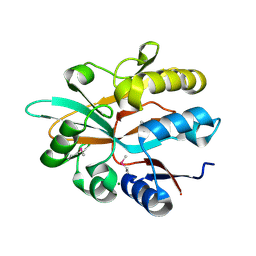 | | Accommodation of massive sequence variation in Nanoarchaeota by the C-type lectin fold | | Descriptor: | AvpA, MAGNESIUM ION | | Authors: | Handa, S, Paul, B, Miller, J, Valentine, D, Ghosh, P. | | Deposit date: | 2016-03-08 | | Release date: | 2016-11-09 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (2.521 Å) | | Cite: | Conservation of the C-type lectin fold for accommodating massive sequence variation in archaeal diversity-generating retroelements.
Bmc Struct.Biol., 16, 2016
|
|
5ELQ
 
 | |
5EM9
 
 | |
5EMB
 
 | |
5EMA
 
 | |
