6I8A
 
 | |
3FUB
 
 | | Crystal structure of GDNF-GFRalpha1 complex | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Parkash, V, Goldman, A. | | Deposit date: | 2009-01-14 | | Release date: | 2009-06-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Comparison of GFL-GFRalpha complexes: further evidence relating GFL bend angle to RET signalling
Acta Crystallogr.,Sect.F, 65, 2009
|
|
6FWK
 
 | |
6G0A
 
 | |
8B67
 
 | |
8B6K
 
 | |
8B7E
 
 | |
8B77
 
 | |
8B76
 
 | |
8B79
 
 | |
2V5E
 
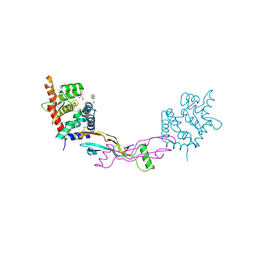 | | The structure of the GDNF:Coreceptor complex: Insights into RET signalling and heparin binding. | | Descriptor: | 1,2-ETHANEDIOL, 1,3,4,6-tetra-O-sulfo-beta-D-fructofuranose-(2-1)-2,3,4,6-tetra-O-sulfonato-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Parkash, V, Leppanen, V.-M, Virtanen, H, Jurvansuu, J.-M, Bespalov, M.M, Sidorova, Y.A, Runeberg-Roos, P, Saarma, M, Goldman, A. | | Deposit date: | 2008-10-03 | | Release date: | 2008-10-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The Structure of the Glial Cell Line-Derived Neurotrophic Factor-Coreceptor Complex: Insights Into Ret Signaling and Heparin Binding.
J.Biol.Chem., 283, 2008
|
|
2W50
 
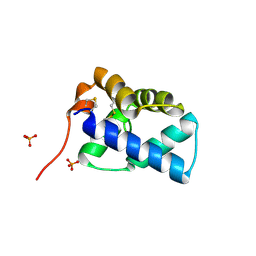 | | N-terminal domain of human conserved dopamine neurotrophic factor (CDNF) | | Descriptor: | ARMET-LIKE PROTEIN 1, PHOSPHATE ION | | Authors: | Parkash, V, Lindholm, P, Peranen, J, Kalkkinen, N, Oksanen, E, Saarma, M, Leppanen, V.M, Goldman, A. | | Deposit date: | 2008-12-03 | | Release date: | 2009-03-17 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Structure of the Conserved Neurotrophic Factors Manf and Cdnf Explains Why They are Bifunctional.
Protein Eng.Des.Sel., 22, 2009
|
|
2W51
 
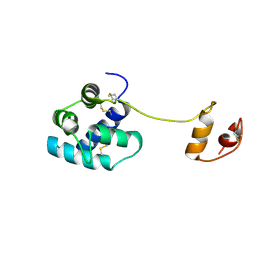 | | Human mesencephalic astrocyte-derived neurotrophic factor (MANF) | | Descriptor: | PROTEIN ARMET | | Authors: | Parkash, V, Lindholm, P, Peranen, J, Kalkkinen, N, Oksanen, E, Saarma, M, Leppanen, V.M, Goldman, A. | | Deposit date: | 2008-12-03 | | Release date: | 2009-03-17 | | Last modified: | 2019-04-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Structure of the Conserved Neurotrophic Factors Manf and Cdnf Explains Why They are Bifunctional.
Protein Eng.Des.Sel., 22, 2009
|
|
6QIB
 
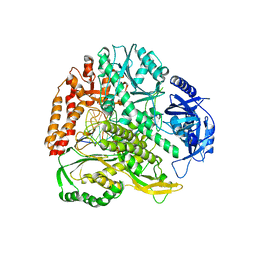 | |
6H1V
 
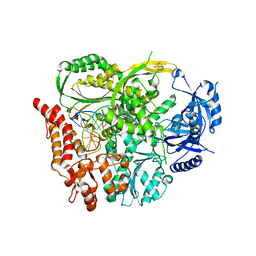 | |
4A0Q
 
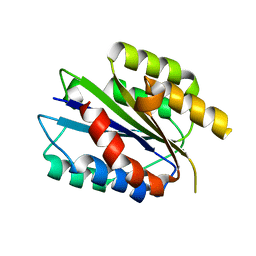 | | Activated Conformation of Integrin alpha1 I-Domain mutant | | Descriptor: | INTEGRIN ALPHA-1, MAGNESIUM ION | | Authors: | Lahti, M, Bligt, E, Niskanen, H, Parkash, V, Brandt, A.-M, Jokinen, J, Patrikainen, P, Kapyla, J, Heino, J, Salminen, T.A. | | Deposit date: | 2011-09-12 | | Release date: | 2011-10-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of Collagen Receptor Integrin Aplha1I Domain Carrying the Activating Mutation E317A.
J.Biol.Chem., 286, 2011
|
|
7R3Y
 
 | | The crystal structure of the V426L variant of Pol2CORE in complex with DNA and an incoming nucleotide | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, CALCIUM ION, DNA Primer, ... | | Authors: | Barbari, S.R, Beach, A.K, Markgren, J.G, Parkash, V, Johansson, E, Shcherbakova, P.V. | | Deposit date: | 2022-02-08 | | Release date: | 2022-07-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Enhanced polymerase activity permits efficient synthesis by cancer-associated DNA polymerase ε variants at low dNTP levels.
Nucleic Acids Res., 50, 2022
|
|
7R3X
 
 | | The crystal structure of the L439V variant of Pol2CORE in complex with DNA and an incoming nucleotide | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, ACETATE ION, CALCIUM ION, ... | | Authors: | Barbari, S.R, Beach, A.K, Markgren, J.G, Parkash, V, Johansson, E, Shcherbakova, P.V. | | Deposit date: | 2022-02-08 | | Release date: | 2022-07-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Enhanced polymerase activity permits efficient synthesis by cancer-associated DNA polymerase ε variants at low dNTP levels.
Nucleic Acids Res., 50, 2022
|
|
2Y74
 
 | | THE CRYSTAL STRUCTURE OF HUMAN SOLUBLE PRIMARY AMINE OXIDASE AOC3 IN THE OFF-COPPER CONFORMATION | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Elovaara, H, Kidron, H, Parkash, V, Nymalm, Y, Bligt, E, Ollikka, P, Smith, D.J, Pihlavisto, M, Salmi, M, Jalkanen, S, Salminen, T.A. | | Deposit date: | 2011-01-28 | | Release date: | 2011-06-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Identification of Two Imidazole Binding Sites and Key Residues for Substrate Specificity in Human Primary Amine Oxidase Aoc3.
Biochemistry, 50, 2011
|
|
2Y73
 
 | | THE NATIVE STRUCTURES OF SOLUBLE HUMAN PRIMARY AMINE OXIDASE AOC3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Elovaara, H, Kidron, H, Parkash, V, Nymalm, Y, Bligt, E, Ollikka, P, Smith, D.J, Pihlavisto, M, Salmi, M, Jalkanen, S, Salminen, T.A. | | Deposit date: | 2011-01-28 | | Release date: | 2011-06-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Identification of Two Imidazole Binding Sites and Key Residues for Substrate Specificity in Human Primary Amine Oxidase Aoc3.
Biochemistry, 50, 2011
|
|
