2VNF
 
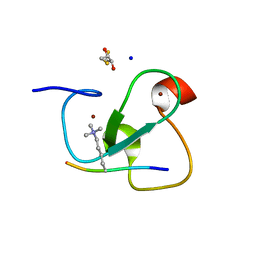 | | MOLECULAR BASIS OF HISTONE H3K4ME3 RECOGNITION BY ING4 | | Descriptor: | (2R,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, HISTONE H3, ... | | Authors: | Palacios, A, Munoz, I.G, Pantoja-Uceda, D, Marcaida, M.J, Torres, D, Martin-Garcia, J.M, Luque, I, Montoya, G, Blanco, F.J. | | Deposit date: | 2008-02-04 | | Release date: | 2008-04-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Molecular Basis of Histone H3K4Me3 Recognition by Ing4
J.Biol.Chem., 283, 2008
|
|
2K1J
 
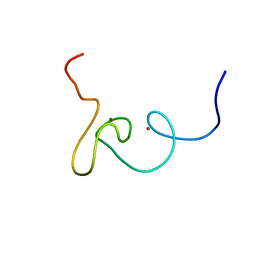 | | Plan homeodomain finger of tumour supressor ING4 | | Descriptor: | Inhibitor of growth protein 4, ZINC ION | | Authors: | Palacios, A, Garcia, P, Padro, D, Lopez-Hernandez, E, Blanco, F.J. | | Deposit date: | 2008-03-05 | | Release date: | 2008-04-15 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure and NMR characterization of the binding to methylated histone tails of the plant homeodomain finger of the tumour suppressor ING4.
Febs Lett., 580, 2006
|
|
4AFL
 
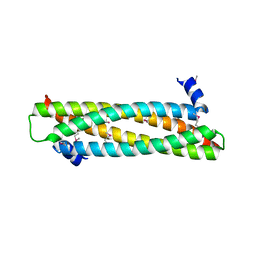 | | The crystal structure of the ING4 dimerization domain reveals the functional organization of the ING family of chromatin binding proteins. | | Descriptor: | INHIBITOR OF GROWTH PROTEIN 4 | | Authors: | Culurgioni, S, Munoz, I.G, Moreno, A, Palacios, A, Villate, M, Palmero, I, Montoya, G, Blanco, F.J. | | Deposit date: | 2012-01-19 | | Release date: | 2012-02-22 | | Last modified: | 2012-07-11 | | Method: | X-RAY DIFFRACTION (2.275 Å) | | Cite: | Crystal Structure of Inhibitor of Growth 4 (Ing4) Dimerization Domain Reveals Functional Organization of Ing Family of Chromatin-Binding Proteins.
J.Biol.Chem., 287, 2012
|
|
6YQ4
 
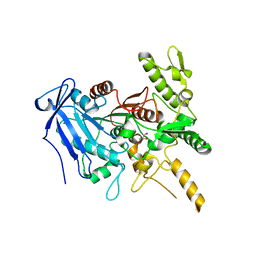 | | Crystal structure of Fusobacterium nucleatum tannase | | Descriptor: | GLYCEROL, MAGNESIUM ION, SPERMIDINE, ... | | Authors: | Mancheno, J.M, Anguita, J, Rodriguez, H. | | Deposit date: | 2020-04-16 | | Release date: | 2021-03-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.399 Å) | | Cite: | A structurally unique Fusobacterium nucleatum tannase provides detoxicant activity against gallotannins and pathogen resistance.
Microb Biotechnol, 15, 2022
|
|
2M1R
 
 | | PHD domain of ING4 N214D mutant | | Descriptor: | Inhibitor of growth protein 4, ZINC ION | | Authors: | Blanco, F.J. | | Deposit date: | 2012-12-05 | | Release date: | 2012-12-26 | | Method: | SOLUTION NMR | | Cite: | Functional impact of cancer-associated mutations in the tumor suppressor protein ING4.
Carcinogenesis, 31, 2010
|
|
6C6I
 
 | | Crystal structure of a chimeric NDM-1 metallo-beta-lactamase harboring the IMP-1 L3 loop | | Descriptor: | Metallo-beta-lactamase type 2 chimera, ZINC ION | | Authors: | Otero, L, Giannini, E, Klinke, S, Palacios, A, Mojica, M, Bonomo, R, Llarrull, L, Vila, A. | | Deposit date: | 2018-01-18 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The Reaction Mechanism of Metallo-beta-Lactamases Is Tuned by the Conformation of an Active-Site Mobile Loop.
Antimicrob. Agents Chemother., 63, 2019
|
|
6CAC
 
 | | Crystal structure of NDM-1 metallo-beta-lactamase harboring an insertion of a Pro residue in L3 loop | | Descriptor: | CADMIUM ION, CALCIUM ION, COBALT (II) ION, ... | | Authors: | Alzari, P.M, Giannini, E, Palacios, A, Mojica, M, Bonomo, R, Llarrull, L, Vila, A. | | Deposit date: | 2018-01-30 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | The Reaction Mechanism of Metallo-beta-Lactamases Is Tuned by the Conformation of an Active-Site Mobile Loop.
Antimicrob. Agents Chemother., 63, 2019
|
|
7K7K
 
 | |
7KGN
 
 | | S. Typhi YcbB - ertapenem complex | | Descriptor: | (4R,5S)-3-({(3S,5S)-5-[(3-carboxyphenyl)carbamoyl]pyrrolidin-3-yl}sulfanyl)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-4-methyl-4,5-dihydro-1H-pyrrole-2-carboxylic acid, L,D-transpeptidase | | Authors: | Caveney, N.A, Strynadka, N.C.J. | | Deposit date: | 2020-10-18 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural and Cellular Insights into the l,d-Transpeptidase YcbB as a Therapeutic Target in Citrobacter rodentium, Salmonella Typhimurium, and Salmonella Typhi Infections.
Antimicrob.Agents Chemother., 65, 2021
|
|
7KGM
 
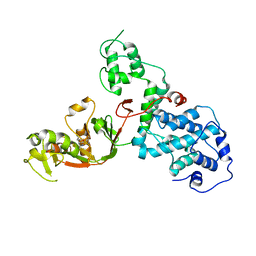 | | C. rodentium YcbB - ertapenem complex | | Descriptor: | (4R,5S)-3-({(3S,5S)-5-[(3-carboxyphenyl)carbamoyl]pyrrolidin-3-yl}sulfanyl)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-4-methyl-4,5-dihydro-1H-pyrrole-2-carboxylic acid, Putative exported protein | | Authors: | Caveney, N.A, Strynadka, N.C.J. | | Deposit date: | 2020-10-17 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and Cellular Insights into the l,d-Transpeptidase YcbB as a Therapeutic Target in Citrobacter rodentium, Salmonella Typhimurium, and Salmonella Typhi Infections.
Antimicrob.Agents Chemother., 65, 2021
|
|
