5YOY
 
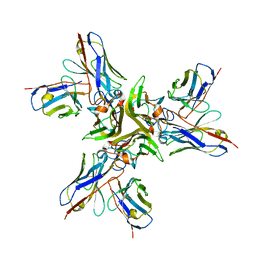 | | Crystal structure of the human tumor necrosis factor in complex with golimumab Fv | | Descriptor: | Golimumab heavy chain variable region, Golimumab light chain variable region, Tumor necrosis factor | | Authors: | Ono, M, Horita, S, Sato, Y, Nomura, Y, Iwata, S, Nomura, N. | | Deposit date: | 2017-10-31 | | Release date: | 2018-05-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.727 Å) | | Cite: | Structural basis for tumor necrosis factor blockade with the therapeutic antibody golimumab
Protein Sci., 27, 2018
|
|
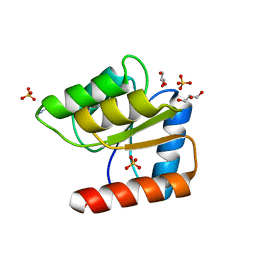
1JBE
 
 | |
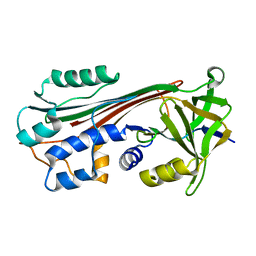
1C8O
 
 | |
2FOT
 
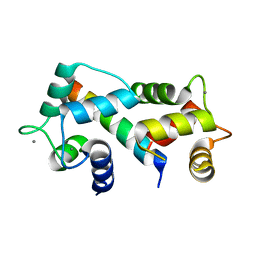 | | Crystal structure of the complex between calmodulin and alphaII-spectrin | | Descriptor: | CALCIUM ION, Calmodulin, alpha-II spectrin Spectrin | | Authors: | Simonovic, M, Zhang, Z, Cianci, C.D, Steitz, T.A, Morrow, J.S. | | Deposit date: | 2006-01-13 | | Release date: | 2006-09-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure of the calmodulin alphaII-spectrin complex provides insight into the regulation of cell plasticity.
J.Biol.Chem., 281, 2006
|
|
1M93
 
 | |
1IMV
 
 | | 2.85 A crystal structure of PEDF | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PIGMENT EPITHELIUM-DERIVED FACTOR | | Authors: | Simonovic, M, Gettins, P.G.W, Volz, K. | | Deposit date: | 2001-05-11 | | Release date: | 2001-09-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal structure of human PEDF, a potent anti-angiogenic and neurite growth-promoting factor.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1J8E
 
 | | Crystal structure of ligand-binding repeat CR7 from LRP | | Descriptor: | CALCIUM ION, LOW-DENSITY LIPOPROTEIN RECEPTOR-RELATED PROTEIN 1 | | Authors: | Simonovic, M, Dolmer, K, Huang, W, Strickland, D.K, Volz, K, Gettins, P.G.W. | | Deposit date: | 2001-05-21 | | Release date: | 2001-12-19 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Calcium coordination and pH dependence of the calcium affinity of ligand-binding repeat CR7 from the LRP. Comparison with related domains from the LRP and the LDL receptor.
Biochemistry, 40, 2001
|
|
3CME
 
 | |
3CMA
 
 | |
7P55
 
 | |
7P5Q
 
 | |
7P5S
 
 | |
7P5G
 
 | |
3EDU
 
 | | Crystal structure of the ankyrin-binding domain of human erythroid spectrin | | Descriptor: | Spectrin beta chain, erythrocyte | | Authors: | Simonovic, M, Stabach, P, Simonovic, I, Steitz, T.A, Morrow, J.S. | | Deposit date: | 2008-09-03 | | Release date: | 2009-02-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structure of the ankyrin-binding site of {beta}-spectrin reveals how tandem spectrin-repeats generate unique ligand-binding properties
Blood, 113, 2009
|
|
5FZO
 
 | | Crystal structure of the catalytic domain of human JmjD1C | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, MANGANESE (II) ION, ... | | Authors: | Nowak, R, Talon, R, Krojer, T, Goubin, S, McDonough, M, Fairhead, M, Oppermann, U, Johansson, C. | | Deposit date: | 2016-03-15 | | Release date: | 2016-03-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystal Structure of the Catalytic Domain of Human Jmjd1C
To be Published
|
|
2KMJ
 
 | | High resolution NMR solution structure of a complex of HIV-2 TAR RNA and a synthetic tripeptide in a 1:2 stoichiometry | | Descriptor: | Pyrimidinylpeptide, RNA (28-MER) | | Authors: | Ferner, J, Suhartono, M, Breitung, S, Jonker, H.R.A, Hennig, M, Woehnert, J, Goebel, M, Schwalbe, H. | | Deposit date: | 2009-07-30 | | Release date: | 2009-08-18 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Structures of HIV TAR RNA-ligand complexes reveal higher binding stoichiometries.
Chembiochem, 10, 2009
|
|
6O2T
 
 | | Acetylated Microtubules | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Eshun-Wilson, L, Zhang, R, Portran, D, Nachury, M.V, Toso, D, Lohr, T, Vendruscolo, M, Bonomi, M, Fraser, J.S, Nogales, E. | | Deposit date: | 2019-02-24 | | Release date: | 2019-05-22 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Effects of alpha-tubulin acetylation on microtubule structure and stability.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6O2Q
 
 | | Acetylated Microtubules | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Eshun-Wilson, L, Zhang, R, Portran, D, Nachury, M.V, Toso, D, Lohr, T, Vendruscolo, M, Bonomi, M, Fraser, J.S, Nogales, E. | | Deposit date: | 2019-02-24 | | Release date: | 2019-05-22 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Effects of alpha-tubulin acetylation on microtubule structure and stability.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6O2S
 
 | | Deacetylated Microtubules | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Eshun-Wilson, L, Zhang, R, Portran, D, Nachury, M.V, Toso, D, Lohr, T, Vendruscolo, M, Bonomi, M, Fraser, J.S, Nogales, E. | | Deposit date: | 2019-02-24 | | Release date: | 2019-05-22 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Effects of alpha-tubulin acetylation on microtubule structure and stability.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6O2R
 
 | | Deacetylated Microtubules | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Eshun-Wilson, L, Zhang, R, Portran, D, Nachury, M.V, Toso, D, Lohr, T, Vendruscolo, M, Bonomi, M, Fraser, J.S, Nogales, E. | | Deposit date: | 2019-02-24 | | Release date: | 2019-05-22 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Effects of alpha-tubulin acetylation on microtubule structure and stability.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
4WYP
 
 | | The crystal structure of the A109G mutant of RNase A in complex with 5'AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Ribonuclease pancreatic | | Authors: | French, R.L, Gagne, D, Doucet, N, Simonovic, M. | | Deposit date: | 2014-11-17 | | Release date: | 2015-11-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.502 Å) | | Cite: | Perturbation of the Conformational Dynamics of an Active-Site Loop Alters Enzyme Activity.
Structure, 23, 2015
|
|
4WYZ
 
 | | The crystal structure of the A109G mutant of RNase A in complex with 3'UMP | | Descriptor: | 3'-URIDINEMONOPHOSPHATE, Ribonuclease pancreatic | | Authors: | French, R.L, Gagne, D, Doucet, N, Simonovic, M. | | Deposit date: | 2014-11-18 | | Release date: | 2015-11-18 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.449 Å) | | Cite: | Perturbation of the Conformational Dynamics of an Active-Site Loop Alters Enzyme Activity.
Structure, 23, 2015
|
|
4WYN
 
 | |
6O0X
 
 | | Conformational states of Cas9-sgRNA-DNA ternary complex in the presence of magnesium | | Descriptor: | 3' product of target strand DNA, 5' product of target strand DNA, CRISPR-associated endonuclease Cas9/Csn1, ... | | Authors: | Zhu, X, Clarke, R, Puppala, A.K, Chittori, S, Merk, A, Merrill, B.J, Simonovic, M, Subramaniam, S. | | Deposit date: | 2019-02-17 | | Release date: | 2019-07-10 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Cryo-EM structures reveal coordinated domain motions that govern DNA cleavage by Cas9.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6O0Z
 
 | | Conformational states of Cas9-sgRNA-DNA ternary complex in the presence of magnesium | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, non-target strand DNA, single guide RNA, ... | | Authors: | Zhu, X, Clarke, R, Puppala, A.K, Chittori, S, Merk, A, Merrill, B.J, Simonovic, M, Subramaniam, S. | | Deposit date: | 2019-02-17 | | Release date: | 2019-07-10 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures reveal coordinated domain motions that govern DNA cleavage by Cas9.
Nat.Struct.Mol.Biol., 26, 2019
|
|
