7L0B
 
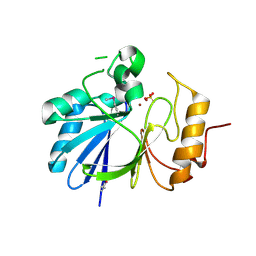 | | Crystal structure of hydroxyacyl glutathione hydrolase (GloB) from Staphylococcus aureus, apoenzyme | | Descriptor: | Hydroxyacylglutathione hydrolase, SULFATE ION, ZINC ION | | Authors: | Miller, J.J, Jez, J.M, Odom John, A.R. | | Deposit date: | 2020-12-11 | | Release date: | 2020-12-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure-guided microbial targeting of antistaphylococcal prodrugs.
Elife, 10, 2021
|
|
7L0A
 
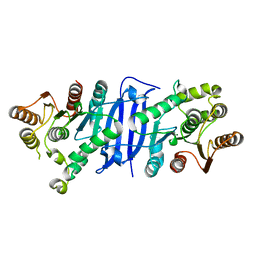 | |
1LDD
 
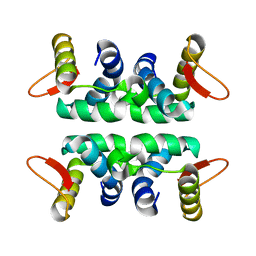 | | Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF Ubiquitin Ligase Complex | | Descriptor: | Anaphase Promoting Complex | | Authors: | Zheng, N, Schulman, B.A, Song, L, Miller, J.J, Jeffrey, P.D, Wang, P, Chu, C, Koepp, D.M, Elledge, S.J, Pagano, M, Conaway, R.C, Conaway, J.W, Harper, J.W, Pavletich, N.P. | | Deposit date: | 2002-04-08 | | Release date: | 2002-05-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex.
Nature, 416, 2002
|
|
1LDK
 
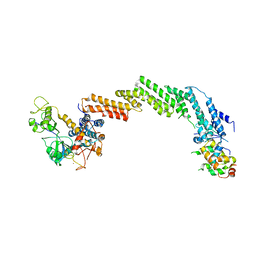 | | Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF Ubiquitin Ligase Complex | | Descriptor: | CULLIN HOMOLOG, CYCLIN A/CDK2-ASSOCIATED PROTEIN P19, SKP2-like protein type gamma, ... | | Authors: | Zheng, N, Schulman, B.A, Song, L, Miller, J.J, Jeffrey, P.D, Wang, P, Chu, C, Koepp, D.M, Elledge, S.J, Pagano, M, Conaway, R.C, Conaway, J.W, Harper, J.W, Pavletich, N.P. | | Deposit date: | 2002-04-08 | | Release date: | 2002-05-08 | | Last modified: | 2019-11-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex.
Nature, 416, 2002
|
|
1LDJ
 
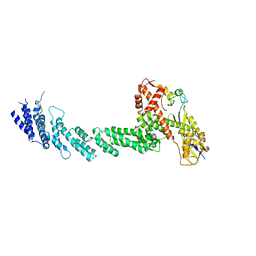 | | Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF Ubiquitin Ligase Complex | | Descriptor: | Cullin homolog 1, ZINC ION, ring-box protein 1 | | Authors: | Zheng, N, Schulman, B.A, Song, L, Miller, J.J, Jeffrey, P.D, Wang, P, Chu, C, Koepp, D.M, Elledge, S.J, Pagano, M, Conaway, R.C, Conaway, J.W, Harper, J.W, Pavletich, N.P. | | Deposit date: | 2002-04-08 | | Release date: | 2002-05-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex.
Nature, 416, 2002
|
|
7T3D
 
 | | CryoEM map of anchor 222-1C06 Fab and lateral patch 2B05 Fab binding H1 HA | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 222-1C06 mAb heavy chain, ... | | Authors: | Han, J, Ward, A.B. | | Deposit date: | 2021-12-07 | | Release date: | 2022-01-12 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Broadly neutralizing antibodies target a haemagglutinin anchor epitope.
Nature, 602, 2022
|
|
7MEM
 
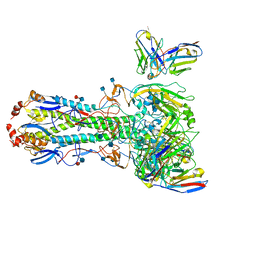 | |
2AST
 
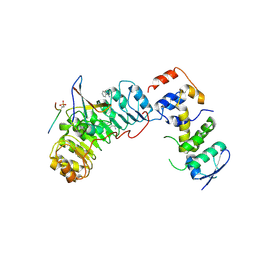 | | Crystal structure of Skp1-Skp2-Cks1 in complex with a p27 peptide | | Descriptor: | BENZAMIDINE, Cyclin-dependent kinase inhibitor 1B, Cyclin-dependent kinases regulatory subunit 1, ... | | Authors: | Hao, B, Zhang, N, Schulman, B.A, Wu, G, Pagano, M, Pavletich, N.P. | | Deposit date: | 2005-08-24 | | Release date: | 2005-10-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis of the Cks1-Dependent Recognition of p27(Kip1) by the SCF(Skp2) Ubiquitin Ligase.
Mol.Cell, 20, 2005
|
|
6MH4
 
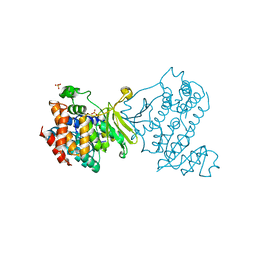 | |
6MH5
 
 | |
2ASS
 
 | | Crystal structure of the Skp1-Skp2-Cks1 complex | | Descriptor: | BENZAMIDINE, Cyclin-dependent kinases regulatory subunit 1, PHOSPHATE ION, ... | | Authors: | Hao, B, Zhang, N, Schulman, B.A, Wu, G, Pagano, M, Pavletich, N.P. | | Deposit date: | 2005-08-24 | | Release date: | 2005-10-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Basis of the Cks1-Dependent Recognition of p27(Kip1) by the SCF(Skp2) Ubiquitin Ligase.
Mol.Cell, 20, 2005
|
|
8G19
 
 | | Crystal structure of monoreactive 4C05 human Fab | | Descriptor: | Heavy chain of anti-influenza human 4C05 Fab fragment, Light chain of anti-influenza human 4C05 Fab fragment | | Authors: | Borowska, M.T, Adams, E.J. | | Deposit date: | 2023-02-01 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Biochemical and biophysical characterization of natural polyreactivity in antibodies.
Cell Rep, 42, 2023
|
|
8G1B
 
 | | Crystal structure of polyreactive 2G02 human Fab | | Descriptor: | The heavy chain of human 2G02 Fab fragment, The light chain of human 2G02 Fab fragment | | Authors: | Borowska, M.T, Adams, E.J. | | Deposit date: | 2023-02-01 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Biochemical and biophysical characterization of natural polyreactivity in antibodies.
Cell Rep, 42, 2023
|
|
8G1C
 
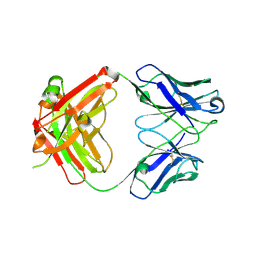 | | Crystal structure of polyreactive 3B03 human Fab | | Descriptor: | Heavy chain of monoreactive 3B03 human Fab fragment, Light chain of monoreactive 3B03 human Fab fragment | | Authors: | Borowska, M.T, Adams, E.J. | | Deposit date: | 2023-02-01 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Biochemical and biophysical characterization of natural polyreactivity in antibodies.
Cell Rep, 42, 2023
|
|
7MYV
 
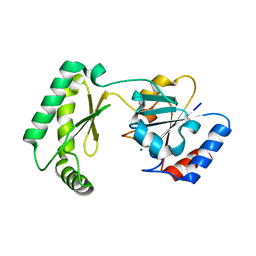 | | Plasmodium falciparum HAD5/PMM | | Descriptor: | MAGNESIUM ION, Phosphomannomutase | | Authors: | Frasse, P.M, Odom John, A.R, Goldberg, D.E. | | Deposit date: | 2021-05-21 | | Release date: | 2022-03-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.51 Å) | | Cite: | Enzymatic and structural characterization of HAD5, an essential phosphomannomutase of malaria-causing parasites.
J.Biol.Chem., 298, 2022
|
|
8FZO
 
 | |
8FZP
 
 | |
