2LHO
 
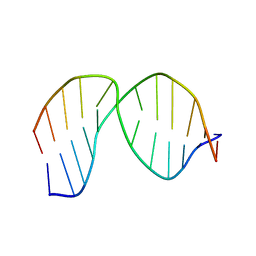 | | Solution Structure of a DNA duplex Containing an Unnatural, Hydrophobic Base Pair | | Descriptor: | DNA (5'-D(*CP*GP*TP*TP*TP*CP*(LHO)P*TP*TP*CP*TP*C)-3'), DNA (5'-D(*GP*AP*GP*AP*AP*(MM7)P*GP*AP*AP*AP*CP*G)-3') | | Authors: | Malyshev, D.A, Pfaff, D.A, Ippoliti, S.L, Hwang, G.T, Dwyer, T.J, Romesberg, F.E. | | Deposit date: | 2011-08-12 | | Release date: | 2012-07-04 | | Method: | SOLUTION NMR | | Cite: | Solution structure, mechanism of replication, and optimization of an unnatural base pair.
Chemistry, 16, 2010
|
|
4C8M
 
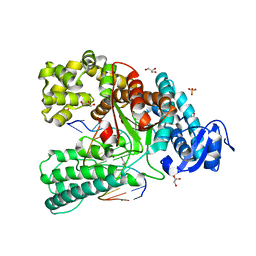 | | Binary complex of the large fragment of DNA polymerase I from Thermus Aquaticus with the aritificial base pair d5SICS-dNaM at the postinsertion site (sequence context 2) | | Descriptor: | GLYCEROL, LARGE FRAGMENT OF TAQ DNA POLYMERASE I, MAGNESIUM ION, ... | | Authors: | Betz, K, Malyshev, D.A, Lavergne, T, Welte, W, Diederichs, K, Romesberg, F.E, Marx, A. | | Deposit date: | 2013-10-01 | | Release date: | 2013-12-11 | | Last modified: | 2017-08-23 | | Method: | X-RAY DIFFRACTION (1.568 Å) | | Cite: | Structural Insights Into DNA Replication without Hydrogen Bonds.
J.Am.Chem.Soc., 135, 2013
|
|
4C8N
 
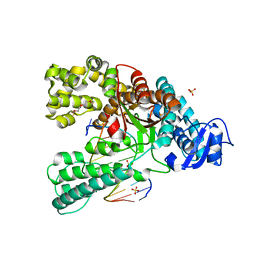 | | Binary complex of the large fragment of DNA polymerase I from Thermus Aquaticus with the aritificial base pair dNaM-d5SICS at the postinsertion site (sequence context 3) | | Descriptor: | LARGE FRAGMENT OF TAQ DNA POLYMERASE I, PRIMER, 5'-D(*AP*CP*CP*AP*CP*GP*GP*CP*GP*CP*LHOP)-3', ... | | Authors: | Betz, K, Malyshev, D.A, Lavergne, T, Welte, W, Diederichs, K, Romesberg, F.E, Marx, A. | | Deposit date: | 2013-10-01 | | Release date: | 2013-12-11 | | Last modified: | 2017-08-23 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structural Insights Into DNA Replication without Hydrogen Bonds.
J.Am.Chem.Soc., 135, 2013
|
|
4CCH
 
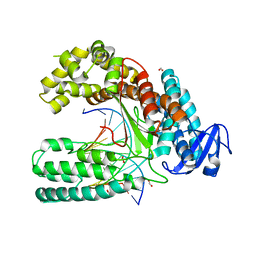 | | Crystal structure of the large fragment of DNA polymerase I from Thermus Aquaticus in an open binary complex with d5SICS as templating nucleotide | | Descriptor: | 5'-D(*AP*AP*CP*LHOP*GP*GP*CP*GP*CP*CP*GP*TP*GP*GP*TP* C)-3', 5'-D(*GP*AP*CP*CP*AP*CP*GP*GP*CP*GP*CP*DOC)-3', DNA POLYMERASE I, ... | | Authors: | Betz, K, Malyshev, D.A, Lavergne, T, Welte, W, Diederichs, K, Romesberg, F.E, Marx, A. | | Deposit date: | 2013-10-23 | | Release date: | 2013-12-11 | | Last modified: | 2013-12-25 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural Insights Into DNA Replication without Hydrogen Bonds.
J.Am.Chem.Soc., 135, 2013
|
|
4C8O
 
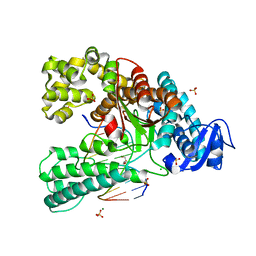 | | Binary complex of the large fragment of DNA polymerase I from Thermus Aquaticus with the aritificial base pair dNaM-d5SICS at the postinsertion site (sequence context 2) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5'-D(*GP*CP*CP*AP*CP*GP*GP*CP*GP*CP*LHOP)-3', 5'-D(*TP*TP*CP*BMNP*GP*CP*GP*CP*CP*GP*TP*GP*GP*CP)-3', ... | | Authors: | Betz, K, Malyshev, D.A, Lavergne, T, Welte, W, Diederichs, K, Romesberg, F.E, Marx, A. | | Deposit date: | 2013-10-01 | | Release date: | 2013-12-11 | | Last modified: | 2017-08-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Insights Into DNA Replication without Hydrogen Bonds.
J.Am.Chem.Soc., 135, 2013
|
|
4C8K
 
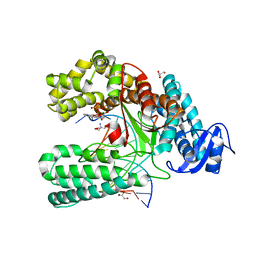 | | Crystal structure of the large fragment of DNA polymerase I from Thermus Aquaticus in a partially closed complex with the artificial base pair d5SICS-dNaMTP | | Descriptor: | ((2R,3S,5R)-3-hydroxy-5-(3-methoxynaphthalen-2-yl)methyl-tetrahydrogen-triphosphate, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5'-D(*AP*AP*C*LHOP*GP*GP*CP*GP*CP*CP*GP*TP*GP*GP*TP*C)-3', ... | | Authors: | Betz, K, Malyshev, D.A, Lavergne, T, Welte, W, Diederichs, K, Romesberg, F.E, Marx, A. | | Deposit date: | 2013-10-01 | | Release date: | 2013-12-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structural Insights Into DNA Replication without Hydrogen Bonds.
J.Am.Chem.Soc., 135, 2013
|
|
4C8L
 
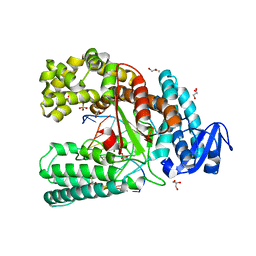 | | Binary complex of the large fragment of DNA polymerase I from Thermus Aquaticus with the artificial base pair dNaM-d5SICS at the postinsertion site (sequence context 1) | | Descriptor: | 5'-D(*AP*CP*CP*AP*CP*GP*GP*CP*GP*CP*LHOP)-3', 5'-D(*AP*GP*BMNP*GP*CP*GP*CP*CP*GP*TP*GP*GP*TP)-3', DNA POLYMERASE I, ... | | Authors: | Betz, K, Malyshev, D.A, Lavergne, T, Welte, W, Diederichs, K, Romesberg, F.E, Marx, A. | | Deposit date: | 2013-10-01 | | Release date: | 2013-12-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Insights Into DNA Replication without Hydrogen Bonds.
J.Am.Chem.Soc., 135, 2013
|
|
3RTV
 
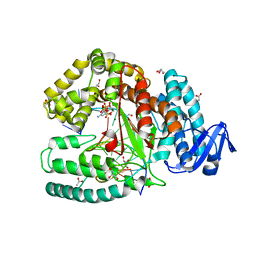 | | Crystal structure of the large fragment of DNA polymerase I from Thermus Aquaticus in a closed ternary complex with natural primer/template DNA | | Descriptor: | (5'-D(*AP*AP*AP*GP*CP*GP*CP*GP*CP*CP*GP*TP*GP*GP*TP*C)-3'), (5'-D(*GP*AP*CP*CP*AP*CP*GP*GP*CP*GP*CP*(DDG))-3'), 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Marx, A, Diederichs, K, Betz, K. | | Deposit date: | 2011-05-04 | | Release date: | 2012-05-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | KlenTaq polymerase replicates unnatural base pairs by inducing a Watson-Crick geometry.
Nat.Chem.Biol., 8, 2012
|
|
3SV4
 
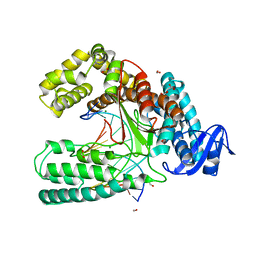 | | Crystal structure of the large fragment of DNA polymerase I from Thermus Aquaticus in an open binary complex with dT as templating nucleobase | | Descriptor: | (5'-D(*AP*AP*AP*TP*GP*GP*CP*GP*CP*CP*GP*TP*GP*GP*TP*C)-3'), (5'-D(*GP*AP*CP*CP*AP*CP*GP*GP*CP*GP*CP*(DOC))-3'), DNA polymerase I, ... | | Authors: | Betz, K, Diederichs, K, Marx, A. | | Deposit date: | 2011-07-12 | | Release date: | 2012-06-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | KlenTaq polymerase replicates unnatural base pairs by inducing a Watson-Crick geometry.
Nat.Chem.Biol., 8, 2012
|
|
3SYZ
 
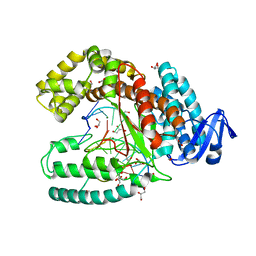 | | Crystal structure of the large fragment of DNA polymerase I from Thermus Aquaticus in an open binary complex with dNaM as templating nucleobase | | Descriptor: | (5'-D(*AP*AP*AP*(BMN)P*GP*GP*CP*GP*CP*CP*GP*TP*GP*GP*TP*C)-3'), (5'-D(*GP*AP*CP*CP*AP*CP*GP*GP*CP*GP*CP*(DOC))-3'), DNA polymerase I, ... | | Authors: | Betz, K, Diederichs, K, Marx, A. | | Deposit date: | 2011-07-18 | | Release date: | 2012-06-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.952 Å) | | Cite: | KlenTaq polymerase replicates unnatural base pairs by inducing a Watson-Crick geometry.
Nat.Chem.Biol., 8, 2012
|
|
3SZ2
 
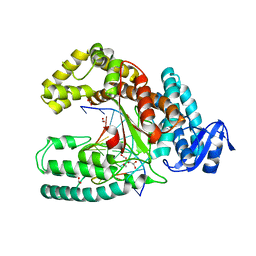 | | Crystal structure of the large fragment of DNA polymerase I from Thermus Aquaticus in an open binary complex with dG as templating nucleobase | | Descriptor: | (5'-D(*AP*AP*AP*GP*CP*GP*CP*GP*CP*CP*GP*TP*GP*GP*TP*C)-3'), (5'-D(*GP*AP*CP*CP*AP*CP*GP*GP*CP*GP*CP*(DDG))-3'), DNA polymerase I, ... | | Authors: | Betz, K, Marx, A, Diederichs, K. | | Deposit date: | 2011-07-18 | | Release date: | 2012-06-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | KlenTaq polymerase replicates unnatural base pairs by inducing a Watson-Crick geometry.
Nat.Chem.Biol., 8, 2012
|
|
3SV3
 
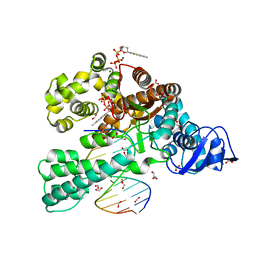 | | Crystal structure of the large fragment of DNA polymerase I from Thermus Aquaticus in a closed ternary complex with the artificial base pair dNaM-d5SICSTP | | Descriptor: | (5'-D(*AP*AP*AP*(BMN)P*GP*GP*CP*GP*CP*CP*GP*TP*GP*GP*TP*C)-3'), (5'-D(*GP*AP*CP*CP*AP*CP*GP*GP*CP*GP*CP*(DOC))-3'), 2-{2-deoxy-5-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]oxy}phosphoryl]-beta-D-erythro-pentofuranosyl}-6-methylisoquinoline-1(2H)-thione, ... | | Authors: | Betz, K, Diederichs, K, Marx, A. | | Deposit date: | 2011-07-12 | | Release date: | 2012-06-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | KlenTaq polymerase replicates unnatural base pairs by inducing a Watson-Crick geometry.
Nat.Chem.Biol., 8, 2012
|
|
