6N9E
 
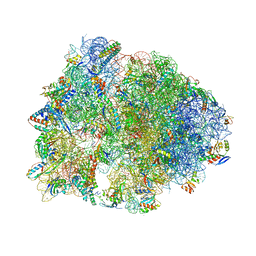 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with a short substrate mimic CC-Pmn and bound to mRNA and P-site tRNA at 3.7A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S Ribosomal Protein S19, ... | | Authors: | Melnikov, S.V, Khabibullina, N.F, Mairhofer, E, Vargas-Rodriguez, O, Reynolds, N.M, Micura, R, Soll, D, Polikanov, Y.S. | | Deposit date: | 2018-12-03 | | Release date: | 2018-12-12 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Mechanistic insights into the slow peptide bond formation with D-amino acids in the ribosomal active site.
Nucleic Acids Res., 47, 2019
|
|
6N9F
 
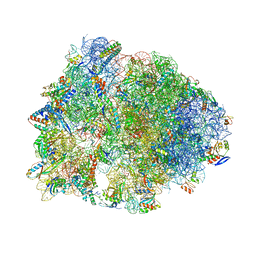 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with a short substrate mimic ACCA-DPhe and bound to mRNA and P-site tRNA at 3.7A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S Ribosomal Protein S19, ... | | Authors: | Melnikov, S.V, Khabibullina, N.F, Mairhofer, E, Vargas-Rodriguez, O, Reynolds, N.M, Micura, R, Soll, D, Polikanov, Y.S. | | Deposit date: | 2018-12-03 | | Release date: | 2018-12-12 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Mechanistic insights into the slow peptide bond formation with D-amino acids in the ribosomal active site.
Nucleic Acids Res., 47, 2019
|
|
7L3R
 
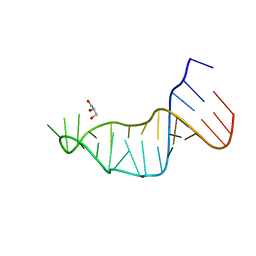 | |
5Y87
 
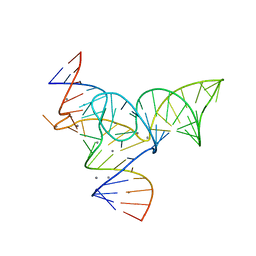 | |
5Y85
 
 | |
5DUN
 
 | | The crystal structure of OMe substituted twister ribozyme | | Descriptor: | MAGNESIUM ION, POTASSIUM ION, RNA (54-MER) | | Authors: | Ren, A, Patel, D.J, Micura, R, Rajashankar, K.R. | | Deposit date: | 2015-09-19 | | Release date: | 2015-10-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | A Mini-Twister Variant and Impact of Residues/Cations on the Phosphodiester Cleavage of this Ribozyme Class.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
6JQ6
 
 | | Hatchet Ribozyme Structure soaking with Ir(NH3)6+ | | Descriptor: | IRIDIUM HEXAMMINE ION, RNA (81-MER) | | Authors: | Ren, A, Zheng, L. | | Deposit date: | 2019-03-29 | | Release date: | 2019-06-12 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.626 Å) | | Cite: | Hatchet ribozyme structure and implications for cleavage mechanism.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6JQ5
 
 | | The structure of Hatchet Ribozyme | | Descriptor: | MAGNESIUM ION, RNA (82-MER) | | Authors: | Ren, A, Zheng, L. | | Deposit date: | 2019-03-29 | | Release date: | 2019-06-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.059 Å) | | Cite: | Hatchet ribozyme structure and implications for cleavage mechanism.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
