8JHF
 
 | | Native SUV420H1 bound to 167-bp nucleosome | | Descriptor: | DNA (160-MER), Histone H2A.Z, Histone H2B type 1-K, ... | | Authors: | Lin, F, Li, W. | | Deposit date: | 2023-05-23 | | Release date: | 2023-11-15 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.68 Å) | | Cite: | Structural basis of nucleosomal H4K20 recognition and methylation by SUV420H1 methyltransferase.
Cell Discov, 9, 2023
|
|
8JHG
 
 | | Native SUV420H1 bound to 167-bp nucleosome | | Descriptor: | DNA (160-MER), Histone H2A type 1-B/E, Histone H2B type 1-K, ... | | Authors: | Lin, F, Li, W. | | Deposit date: | 2023-05-23 | | Release date: | 2023-11-22 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Structural basis of nucleosomal H4K20 recognition and methylation by SUV420H1 methyltransferase.
Cell Discov, 9, 2023
|
|
6FCE
 
 | | NMR ensemble of Macrocyclic Peptidomimetic Containing Constrained a,a-dialkylated Amino Acids with Potent and Selective Activity at Human Melanocortin Receptors | | Descriptor: | ACP-HIS-DPHE-ARG-TRP-ASP-NH2 | | Authors: | Brancaccio, D, Carotenuto, A, Grieco, P, Merlino, F, Zhou, Y, Cai, M, Yousif, A.M, Di Maro, S, Novellino, E, Hruby, V.J. | | Deposit date: | 2017-12-20 | | Release date: | 2018-04-25 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Development of Macrocyclic Peptidomimetics Containing Constrained alpha , alpha-Dialkylated Amino Acids with Potent and Selective Activity at Human Melanocortin Receptors.
J. Med. Chem., 61, 2018
|
|
6WY6
 
 | | Crystal structure of S. cerevisiae Atg8 in complex with Ede1 (1220-1247) | | Descriptor: | Autophagy-related protein 8, EH domain-containing and endocytosis protein 1 | | Authors: | Zheng, Y, Wilfling, F, Baumeister, W, Schulman, B.A. | | Deposit date: | 2020-05-12 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.773 Å) | | Cite: | A Selective Autophagy Pathway for Phase-Separated Endocytic Protein Deposits.
Mol.Cell, 80, 2020
|
|
6HVC
 
 | | NMR structure of Urotensin Peptide Asp-c[Cys-Phe-Trp-(N-Me)Lys-Tyr-Cys]-Val in SDS solution | | Descriptor: | Urotensin-2 | | Authors: | Brancaccio, D, Carotenuto, A, Merlino, F, Billard, E, Yousif, A.M, Di Maro, S, Abate, L, Bellavita, R, D'Emmanuele di Villa Bianca, R, Santicioli, P, Marinelli, L, Novellino, E, Hebert, T.E, Lubell, W.D, Chatenet, D, Grieco, P. | | Deposit date: | 2018-10-10 | | Release date: | 2019-01-16 | | Last modified: | 2025-04-09 | | Method: | SOLUTION NMR | | Cite: | Functional Selectivity Revealed by N-Methylation Scanning of Human Urotensin II and Related Peptides.
J.Med.Chem., 62, 2019
|
|
6HVB
 
 | | NMR structure of Urotensin Peptide Asp-c[Cys-Phe-(N-Me)Trp-Lys-Tyr-Cys]-Val in SDS solution | | Descriptor: | Urotensin-2 | | Authors: | Brancaccio, D, Carotenuto, A, Merlino, F, Billard, E, Yousif, A.M, Di Maro, S, Abate, L, Bellavita, R, D'Emmanuele di Villa Bianca, R, Santicioli, P, Marinelli, L, Novellino, E, Hebert, T.E, Lubell, W.D, Chatenet, D, Grieco, P. | | Deposit date: | 2018-10-10 | | Release date: | 2019-01-16 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Functional Selectivity Revealed by N-Methylation Scanning of Human Urotensin II and Related Peptides.
J.Med.Chem., 62, 2019
|
|
7ZBQ
 
 | | Structure of the ADP-ribosyltransferase TccC3HVR from Photorhabdus Luminescens | | Descriptor: | TccC3 | | Authors: | Lindemann, F, Belyy, A, Friedrich, D, Schmieder, P, Bardiaux, B, Roderer, D, Funk, J, Protze, J, Bieling, P, Oschkinat, H, Raunser, S. | | Deposit date: | 2022-03-24 | | Release date: | 2022-06-29 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Mechanism of threonine ADP-ribosylation of F-actin by a Tc toxin.
Nat Commun, 13, 2022
|
|
2QHX
 
 | | Structure of Pteridine Reductase from Leishmania major complexed with a ligand | | Descriptor: | IODIDE ION, METHYL 1-(4-{[(2,4-DIAMINOPTERIDIN-6-YL)METHYL](METHYL)AMINO}BENZOYL)PIPERIDINE-4-CARBOXYLATE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Gibellini, F, Mcluskey, K, Tulloch, L, Hunter, W.N. | | Deposit date: | 2007-07-03 | | Release date: | 2007-12-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Discovery of potent pteridine reductase inhibitors to guide antiparasite drug development.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
9QE1
 
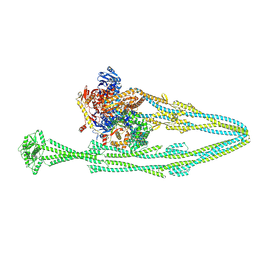 | |
9QE0
 
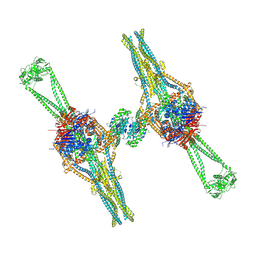 | |
4UV3
 
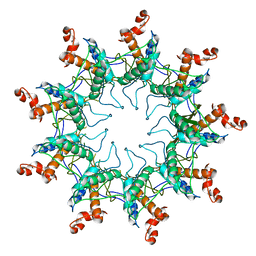 | | Structure of the curli transport lipoprotein CsgG in its membrane- bound conformation | | Descriptor: | CURLI PRODUCTION ASSEMBLY/TRANSPORT COMPONENT CSGG | | Authors: | Goyal, P, Krasteva, P.V, Gerven, N.V, Gubellini, F, Broeck, I.V.D, Troupiotis-Tsailaki, A, Jonckheere, W, Pehau-Arnaudet, G, Pinkner, J.S, Chapman, M.R, Hultgren, S.J, Howorka, S, Fronzes, R, Remaut, H. | | Deposit date: | 2014-08-04 | | Release date: | 2014-09-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.59 Å) | | Cite: | Structural and Mechanistic Insights Into the Bacterial Amyloid Secretion Channel Csgg.
Nature, 516, 2014
|
|
4UV2
 
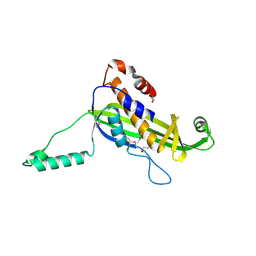 | | Structure of the curli transport lipoprotein CsgG in a non-lipidated, pre-pore conformation | | Descriptor: | CURLI PRODUCTION TRANSPORT COMPONENT CSGG | | Authors: | Goyal, P, Krasteva, P.V, Gerven, N.V, Gubellini, F, Broeck, I.V.D, Troupiotis-Tsailaki, A, Jonckheere, W, Pehau-Arnaudet, G, Pinkner, J.S, Chapman, M.R, Hultgren, S.J, Howorka, S, Fronzes, R, Remaut, H. | | Deposit date: | 2014-08-04 | | Release date: | 2014-09-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and Mechanistic Insights Into the Bacterial Amyloid Secretion Channel Csgg.
Nature, 516, 2014
|
|
1UG3
 
 | | C-terminal portion of human eIF4GI | | Descriptor: | eukaryotic protein synthesis initiation factor 4G | | Authors: | Bellsolell, L, Cho-Park, P.F, Poulin, F, Sonenberg, N, Burley, S.K. | | Deposit date: | 2003-06-11 | | Release date: | 2005-03-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Two structurally atypical HEAT domains in the C-terminal portion of human eIF4G support binding to eIF4A and Mnk1
Structure, 14, 2006
|
|
8BFN
 
 | | E. coli Wadjet JetABC dimer of dimers | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, JetA, JetB, ... | | Authors: | Roisne-Hamelin, F, Beckert, B, Myasnikov, A, Li, Y, Gruber, S. | | Deposit date: | 2022-10-26 | | Release date: | 2022-12-14 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | DNA-measuring Wadjet SMC ATPases restrict smaller circular plasmids by DNA cleavage.
Mol.Cell, 82, 2022
|
|
8GY0
 
 | | Agrocybe pediades linalool sunthase (Ap.LS) | | Descriptor: | Terpene synthase | | Authors: | Rehka, T, Sharma, D, Lin, F, Lim, C, Choong, Y.K, Chacko, J, Zhang, C. | | Deposit date: | 2022-09-21 | | Release date: | 2023-04-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.988 Å) | | Cite: | Structural Understanding of Fungal Terpene Synthases for the Formation of Linear or Cyclic Terpene Products.
Acs Catalysis, 13, 2023
|
|
3IYP
 
 | | The Interaction of Decay-accelerating Factor with Echovirus 7 | | Descriptor: | Capsid protein, Complement decay-accelerating factor, LAURIC ACID, ... | | Authors: | Plevka, P, Hafenstein, S, Zhang, Y, Harris, K.G, Cifuente, J.O, Bowman, V.D, Chipman, P.R, Lin, F, Medof, D.E, Bator, C.M, Rossmann, M.G. | | Deposit date: | 2010-04-07 | | Release date: | 2010-11-24 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (7.2 Å) | | Cite: | Interaction of decay-accelerating factor with echovirus 7.
J.Virol., 84, 2010
|
|
1NWV
 
 | | SOLUTION STRUCTURE OF A FUNCTIONALLY ACTIVE COMPONENT OF DECAY ACCELERATING FACTOR | | Descriptor: | Complement decay-accelerating factor | | Authors: | Uhrinova, S, Lin, F, Ball, G, Bromek, K, Uhrin, D, Medof, M.E, Barlow, P.N. | | Deposit date: | 2003-02-07 | | Release date: | 2003-04-22 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a functionally active fragment of decay-accelerating factor
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1M11
 
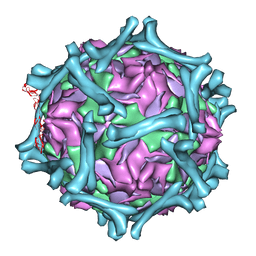 | | structural model of human decay-accelerating factor bound to echovirus 7 from cryo-electron microscopy | | Descriptor: | COAT PROTEIN VP1, COAT PROTEIN VP2, COAT PROTEIN VP3, ... | | Authors: | He, Y, Lin, F, Chipman, P.R, Bator, C.M, Baker, T.S, Shoham, M, Kuhn, R.J, Medof, M.E, Rossmann, M.G. | | Deposit date: | 2002-06-17 | | Release date: | 2002-08-28 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (16 Å) | | Cite: | Structure of decay-accelerating factor bound to echovirus 7: a virus-receptor complex.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
2QZH
 
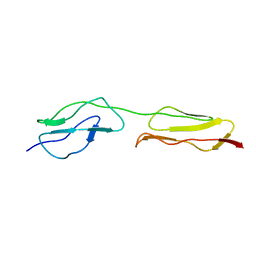 | | SCR2/3 of DAF from the NMR structure 1nwv fitted into a cryoEM reconstruction of CVB3-RD complexed with DAF | | Descriptor: | Complement decay-accelerating factor | | Authors: | Hafenstein, S, Bowman, V.D, Chipman, P.R, Bator Kelly, C.M, Lin, F, Medof, M.E, Rossmann, M.G. | | Deposit date: | 2007-08-16 | | Release date: | 2007-10-30 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (14 Å) | | Cite: | Interaction of decay-accelerating factor with coxsackievirus b3.
J.Virol., 81, 2007
|
|
2QZF
 
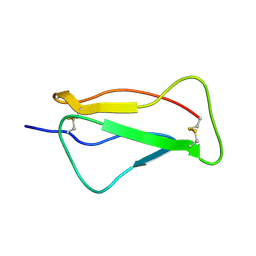 | | SCR1 of DAF from 1ojv fitted into cryoEM density | | Descriptor: | Complement decay-accelerating factor | | Authors: | Hafenstein, S, Bowman, V.D, Chipman, P.R, Bator Kelly, C.M, Lin, F, Medof, M.E, Rossmann, M.G. | | Deposit date: | 2007-08-16 | | Release date: | 2007-10-30 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (14 Å) | | Cite: | Interaction of decay-accelerating factor with coxsackievirus b3.
J.Virol., 81, 2007
|
|
2QZD
 
 | | Fitted structure of SCR4 of DAF into cryoEM density | | Descriptor: | Complement decay-accelerating factor | | Authors: | Hafenstein, S, Bowman, V.D, Chipman, P.R, Bator Kelly, C.M, Lin, F, Medof, M.E, Rossmann, M.G. | | Deposit date: | 2007-08-16 | | Release date: | 2007-10-30 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (14 Å) | | Cite: | Interaction of decay-accelerating factor with coxsackievirus b3.
J.Virol., 81, 2007
|
|
2AIZ
 
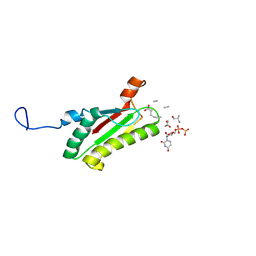 | | Solution structure of peptidoglycan associated lipoprotein from Haemophilus influenza bound to UDP-N-acetylmuramoyl-L-alanyl-D-glutamyl-meso-2,6-diaminopimeloyl-D-alanyl-D-alanine | | Descriptor: | L-alanyl-D-glutamyl-meso-2,6-diaminopimeloyl-D-alanyl-D-alanine, N-acetyl-beta-muramic acid, Outer membrane protein P6 (Fragment), ... | | Authors: | Parsons, L.M, Lin, F, Orban, J, Structure 2 Function Project (S2F) | | Deposit date: | 2005-08-01 | | Release date: | 2006-03-14 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Peptidoglycan recognition by pal, an outer membrane lipoprotein.
Biochemistry, 45, 2006
|
|
1YZ3
 
 | | Structure of human pnmt complexed with cofactor product adohcy and inhibitor SK&F 64139 | | Descriptor: | 7,8-DICHLORO-1,2,3,4-TETRAHYDROISOQUINOLINE, Phenylethanolamine N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Wu, Q, Gee, C.L, Lin, F, Martin, J.L, Grunewald, G.L, McLeish, M.J. | | Deposit date: | 2005-02-27 | | Release date: | 2006-02-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural, mutagenic, and kinetic analysis of the binding of substrates and inhibitors of human phenylethanolamine N-methyltransferase
J.Med.Chem., 48, 2005
|
|
2CH0
 
 | | Solution structure of the human MAN1 C-terminal domain (residues 655- 775) | | Descriptor: | INNER NUCLEAR MEMBRANE PROTEIN MAN1 | | Authors: | Caputo, S, Couprie, J, Duband-Goulet, I, Lin, F, Braud, S, Gondry, M, Worman, H.J, Gilquin, B, Zinn-Justin, S. | | Deposit date: | 2006-03-10 | | Release date: | 2006-05-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The carboxyl-terminal nucleoplasmic region of MAN1 exhibits a DNA binding winged helix domain.
J. Biol. Chem., 281, 2006
|
|
6Z0F
 
 | | Crystal structure of the membrane pseudokinase YukC/EssB from Bacillus subtilis T7SS | | Descriptor: | ESX secretion system protein YukC | | Authors: | Tassinari, M, Bellinzoni, M, Alzari, P.M, Fronzes, R, Gubellini, F. | | Deposit date: | 2020-05-08 | | Release date: | 2021-05-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.553 Å) | | Cite: | The Antibacterial Type VII Secretion System of Bacillus subtilis: Structure and Interactions of the Pseudokinase YukC/EssB.
Mbio, 13, 2022
|
|
