3RVG
 
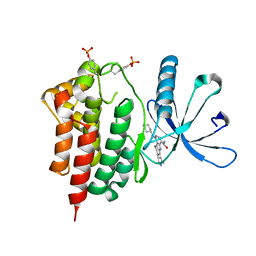 | | Crystals structure of Jak2 with a 1-amino-5H-pyrido[4,3-b]indol-4-carboxamide inhibitor | | Descriptor: | 1-(cyclohexylamino)-7-(1-methyl-1H-pyrazol-4-yl)-5H-pyrido[4,3-b]indole-4-carboxamide, Tyrosine-protein kinase JAK2 | | Authors: | Lim, J, Taoka, B, Otte, R.D, Spencer, K, Dinsmore, C.J, Altman, M.D, Chan, G, Rosenstein, C, Sharma, S, Su, H.P, Szewczak, A.A, Xu, L, Yin, H, Zugay-Murphy, J, Marshall, C.G, Young, J.R. | | Deposit date: | 2011-05-06 | | Release date: | 2012-03-21 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.498 Å) | | Cite: | Discovery of 1-amino-5H-pyrido[4,3-b]indol-4-carboxamide inhibitors of Janus kinase 2 (JAK2) for the treatment of myeloproliferative disorders.
J.Med.Chem., 54, 2011
|
|
2L9F
 
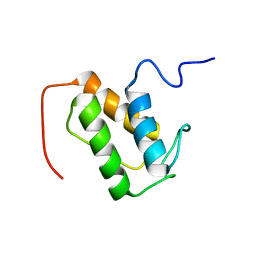 | | NMR solution structure of meACP | | Descriptor: | CalE8 | | Authors: | Lim, J, Yang, D, Liang, Z.X, Kong, R, Murugan, E, Ho, C.L. | | Deposit date: | 2011-02-08 | | Release date: | 2011-06-15 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the acyl carrier protein domain from the highly reducing type I iterative polyketide synthase CalE8
Plos One, 6, 2011
|
|
2LUT
 
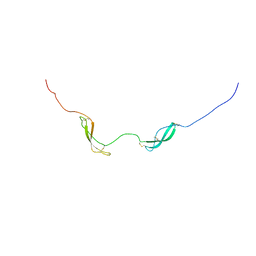 | | NMR solution structure of midkine-a | | Descriptor: | Midkine-related growth factor | | Authors: | Lim, J, Yang, D, Meng, D. | | Deposit date: | 2012-06-21 | | Release date: | 2013-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural and Functional Characterization of Two Zebrafish Midkine Proteins Reveals Importance of the Conserved Hinge for Heparin Binding and Embryogenesis
To be Published
|
|
5KE0
 
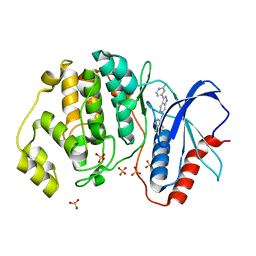 | | Discovery of 1-1H-Pyrazolo 4,3-c pyridine-6-yl urea Inhibitors of Extracellular Signal Regulated Kinase ERK for the Treatment of Cancers | | Descriptor: | 1-[3-(2-methylpyridin-4-yl)-1~{H}-pyrazolo[4,3-c]pyridin-6-yl]-3-(phenylmethyl)urea, Mitogen-activated protein kinase 1, SULFATE ION | | Authors: | Hruza, A, Lim, J. | | Deposit date: | 2016-06-09 | | Release date: | 2016-07-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Discovery of 1-(1H-Pyrazolo[4,3-c]pyridin-6-yl)urea Inhibitors of Extracellular Signal-Regulated Kinase (ERK) for the Treatment of Cancers.
J.Med.Chem., 59, 2016
|
|
2LUU
 
 | | NMR solution structure of midkine-b, mdkb | | Descriptor: | Midkine-related growth factor Mdk2 | | Authors: | Yang, D, Lim, J, Meng, D. | | Deposit date: | 2012-06-21 | | Release date: | 2013-05-01 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure-function analysis of full-length midkine reveals novel residues important for heparin binding and zebrafish embryogenesis.
Biochem.J., 451, 2013
|
|
2M6M
 
 | |
7RSP
 
 | | Structure of the VPS34 kinase domain with compound 14 | | Descriptor: | (7R,8R)-2-[(3R)-3-methylmorpholin-4-yl]-7-(propan-2-yl)-6,7-dihydropyrazolo[1,5-a]pyrazin-4(5H)-one, GLYCEROL, Phosphatidylinositol 3-kinase catalytic subunit type 3 | | Authors: | Hu, D.X, Patel, S, Chen, H, Wang, S, Staben, S, Dimitrova, Y.N, Wallweber, H.A, Lee, J.Y, Chan, G.K.Y, Sneeringer, C.J, Prangley, M.S, Moffat, J.G, Wu, C, Schutt, L.K, Salphati, L, Pang, J, McNamara, E, Huang, H, Chen, Y, Wang, Y, Zhao, W, Lim, J, Murthy, A, Siu, M. | | Deposit date: | 2021-08-11 | | Release date: | 2021-11-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structure-Based Design of Potent, Selective, and Orally Bioavailable VPS34 Kinase Inhibitors.
J.Med.Chem., 65, 2022
|
|
7RSV
 
 | | Structure of the VPS34 kinase domain with compound 5 | | Descriptor: | (5aS,8aR,9S)-2-[(3R)-3-methylmorpholin-4-yl]-5,5a,6,7,8,8a-hexahydro-4H-cyclopenta[e]pyrazolo[1,5-a]pyrazin-4-one, GLYCEROL, Phosphatidylinositol 3-kinase catalytic subunit type 3, ... | | Authors: | Hu, D.X, Patel, S, Chen, H, Wang, S, Staben, S, Dimitrova, Y.N, Wallweber, H.A, Lee, J.Y, Chan, G.K.Y, Sneeringer, C.J, Prangley, M.S, Moffat, J.G, Wu, C, Schutt, L.K, Salphati, L, Pang, J, McNamara, E, Huang, H, Chen, Y, Wang, Y, Zhao, W, Lim, J, Murthy, A, Siu, M. | | Deposit date: | 2021-08-11 | | Release date: | 2021-11-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structure-Based Design of Potent, Selective, and Orally Bioavailable VPS34 Kinase Inhibitors.
J.Med.Chem., 65, 2022
|
|
7RSJ
 
 | | Structure of the VPS34 kinase domain with compound 14 | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, N-{4-[(7R,8R)-4-oxo-7-(propan-2-yl)-4,5,6,7-tetrahydropyrazolo[1,5-a]pyrazin-2-yl]pyridin-2-yl}cyclopropanecarboxamide, ... | | Authors: | Hu, D.X, Patel, S, Chen, H, Wang, S, Staben, S, Dimitrova, Y.N, Wallweber, H.A, Lee, J.Y, Chan, G.K.Y, Sneeringer, C.J, Prangley, M.S, Moffat, J.G, Wu, C, Schutt, L.K, Salphati, L, Pang, J, McNamara, E, Huang, H, Chen, Y, Wang, Y, Zhao, W, Lim, J, Murthy, A, Siu, M. | | Deposit date: | 2021-08-11 | | Release date: | 2021-11-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.881 Å) | | Cite: | Structure-Based Design of Potent, Selective, and Orally Bioavailable VPS34 Kinase Inhibitors.
J.Med.Chem., 65, 2022
|
|
2QJ6
 
 | | Crystal structure analysis of a 14 repeat C-terminal fragment of toxin TcdA in Clostridium difficile | | Descriptor: | Toxin A | | Authors: | Albesa-Jove, D, Bertrand, T, Carpenter, L, Lim, J, Brown, K.A, Fairweather, N. | | Deposit date: | 2007-07-06 | | Release date: | 2008-07-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Solution and crystal structures of the cell binding domain of toxins TcdA and TcdB from Clostridium difficile
To be Published
|
|
4Y73
 
 | | Crystal structure of IRAK4 kinase domain with inhibitor | | Descriptor: | 5-{[(1R,2S)-2-aminocyclohexyl]amino}-N-[1-methyl-3-(trifluoromethyl)-1H-pyrazol-4-yl]pyrazolo[1,5-a]pyrimidine-3-carboxamide, Interleukin-1 receptor-associated kinase 4 | | Authors: | Lesburg, C.A. | | Deposit date: | 2015-02-13 | | Release date: | 2015-05-20 | | Last modified: | 2015-07-15 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Discovery of 5-Amino-N-(1H-pyrazol-4-yl)pyrazolo[1,5-a]pyrimidine-3-carboxamide Inhibitors of IRAK4.
Acs Med.Chem.Lett., 6, 2015
|
|
4WYS
 
 | | Crystal structure of thiolase from Escherichia coli | | Descriptor: | Acetyl-CoA acetyltransferase | | Authors: | Kim, S, Ha, S.C, Ahn, J.W, Kim, E.J, Lim, J.H, Kim, K.J. | | Deposit date: | 2014-11-18 | | Release date: | 2015-10-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Redox-switch regulatory mechanism of thiolase from Clostridium acetobutylicum
Nat Commun, 6, 2015
|
|
7MHC
 
 | | Structure of human STING in complex with MK-1454 | | Descriptor: | (2R,5R,7R,8S,10R,12aR,14R,15S,15aR,16R)-7-(2-amino-6-oxo-1,6-dihydro-9H-purin-9-yl)-14-(6-amino-9H-purin-9-yl)-15,16-difluoro-2,10-bis(sulfanyl)octahydro-2H,10H,12H-5,8-methano-2lambda~5~,10lambda~5~-furo[3,2-l][1,3,6,9,11,2,10]pentaoxadiphosphacyclotetradecine-2,10-dione, Stimulator of interferon genes protein | | Authors: | Lesburg, C.A. | | Deposit date: | 2021-04-15 | | Release date: | 2022-04-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | A kinase-cGAS cascade to synthesize a therapeutic STING activator.
Nature, 603, 2022
|
|
8UQV
 
 | |
8UZH
 
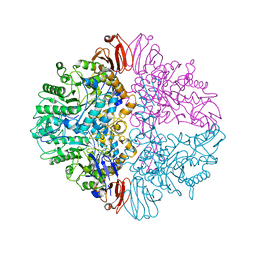 | |
4WYR
 
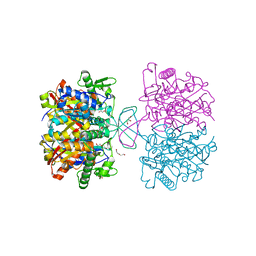 | | Crystal structure of thiolase mutation (V77Q,N153Y,A286K) from Clostridium acetobutylicum | | Descriptor: | Acetyl-CoA acetyltransferase, DI(HYDROXYETHYL)ETHER, GLYCEROL | | Authors: | Kim, S, Ha, S.C, Ahn, J.W, Kim, E.J, Lim, J.H, Kim, K.J. | | Deposit date: | 2014-11-18 | | Release date: | 2015-10-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Redox-switch regulatory mechanism of thiolase from Clostridium acetobutylicum
Nat Commun, 6, 2015
|
|
4XL4
 
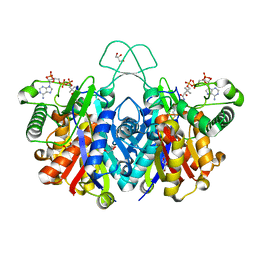 | | Crystal structure of thiolase from Clostridium acetobutylicum in complex with CoA | | Descriptor: | Acetyl-CoA acetyltransferase, COENZYME A, GLYCEROL | | Authors: | Kim, S, Ha, S.C, Ahn, J.W, Kim, E.J, Lim, J.H, Kim, K.J. | | Deposit date: | 2015-01-13 | | Release date: | 2015-10-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Redox-switch regulatory mechanism of thiolase from Clostridium acetobutylicum
Nat Commun, 6, 2015
|
|
4XL2
 
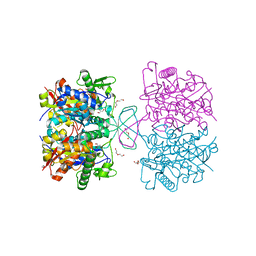 | | Crystal structure of oxidized form of thiolase from Clostridium acetobutylicum | | Descriptor: | ACETATE ION, Acetyl-CoA acetyltransferase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kim, S, Ha, S.C, Ahn, J.W, Kim, E.J, Lim, J.H, Kim, K.J. | | Deposit date: | 2015-01-13 | | Release date: | 2015-10-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Redox-switch regulatory mechanism of thiolase from Clostridium acetobutylicum
Nat Commun, 6, 2015
|
|
4XL3
 
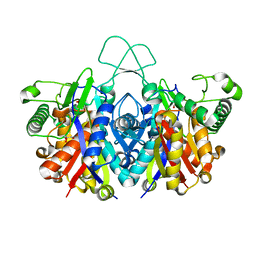 | | Crystal structure of reduced form of thiolase from Clostridium acetobutylicum | | Descriptor: | Acetyl-CoA acetyltransferase, GLYCEROL | | Authors: | Kim, S, Ha, S.C, Ahn, J.W, Kim, E.J, Lim, J.H, Kim, K.J. | | Deposit date: | 2015-01-13 | | Release date: | 2015-10-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Redox-switch regulatory mechanism of thiolase from Clostridium acetobutylicum
Nat Commun, 6, 2015
|
|
5T1S
 
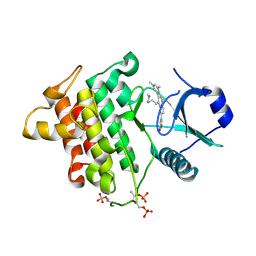 | | Irak4 kinase - compound 1 co-structure | | Descriptor: | 5-[3-(3,5-dimethylphenyl)-4-[4-(methylamino)butyl]quinolin-6-yl]pyridin-3-ol, Interleukin-1 receptor-associated kinase 4 | | Authors: | Fischmann, T.O. | | Deposit date: | 2016-08-22 | | Release date: | 2017-05-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification of quinazoline based inhibitors of IRAK4 for the treatment of inflammation.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
5T1T
 
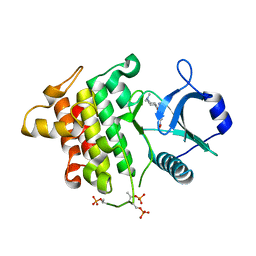 | | Irak4 kinase - compound 1 co-structure | | Descriptor: | Interleukin-1 receptor-associated kinase 4, ~{N},~{N}-dimethyl-4-(6-nitroquinazolin-4-yl)oxy-cyclohexan-1-amine | | Authors: | Fischmann, T.O. | | Deposit date: | 2016-08-22 | | Release date: | 2017-05-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Identification of quinazoline based inhibitors of IRAK4 for the treatment of inflammation.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
1OZ1
 
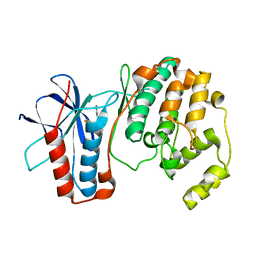 | | P38 MITOGEN-ACTIVATED KINASE IN COMPLEX WITH 4-AZAINDOLE INHIBITOR | | Descriptor: | 3-(4-FLUOROPHENYL)-2-PYRIDIN-4-YL-1H-PYRROLO[3,2-B]PYRIDIN-1-OL, Mitogen-activated protein kinase 14 | | Authors: | Lovejoy, B, Villasenor, A, Browner, M, Dunten, P. | | Deposit date: | 2003-04-07 | | Release date: | 2003-09-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Design and synthesis of 4-azaindoles as inhibitors of p38 MAP kinase.
J.Med.Chem., 46, 2003
|
|
3BWO
 
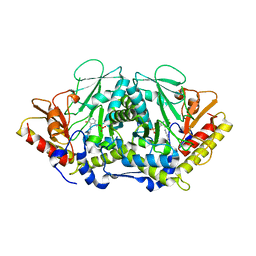 | | L-tryptophan aminotransferase | | Descriptor: | L-tryptophan aminotransferase | | Authors: | Ferrer, J.-L, Noel, J.P, Pojer, F, Bowman, M, Chory, J, Tao, Y. | | Deposit date: | 2008-01-10 | | Release date: | 2009-03-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for the shade avoidance response of plants
To be Published
|
|
3BWN
 
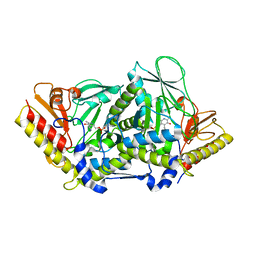 | | L-tryptophan aminotransferase | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, L-tryptophan aminotransferase, PHENYLALANINE, ... | | Authors: | Ferrer, J.-L, Noel, J.P, Pojer, F, Bowman, M, Chory, J, Tao, Y. | | Deposit date: | 2008-01-10 | | Release date: | 2008-04-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants
Cell(Cambridge,Mass.), 133, 2008
|
|
6SLM
 
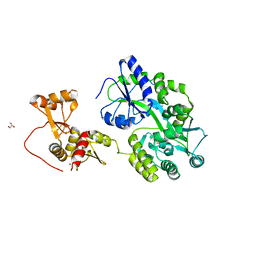 | | Crystal structure of full-length HPV31 E6 oncoprotein in complex with LXXLL peptide of ubiquitin ligase E6AP | | Descriptor: | GLYCEROL, Maltose/maltodextrin-binding periplasmic protein,Protein E6,Ubiquitin-protein ligase E3A, ZINC ION, ... | | Authors: | Conrady, M, Gogl, G, Cousido-Siah, A, Mitschler, A, Trave, G, Simon, C. | | Deposit date: | 2019-08-20 | | Release date: | 2020-09-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of High-Risk Papillomavirus 31 E6 Oncogenic Protein and Characterization of E6/E6AP/p53 Complex Formation.
J.Virol., 95, 2020
|
|
