3D38
 
 | | Crystal structure of new trigonal form of photosynthetic reaction center from Blastochloris viridis. Crystals grown in microfluidics by detergent capture. | | Descriptor: | 15-cis-1,2-dihydroneurosporene, BACTERIOCHLOROPHYLL B, BACTERIOPHEOPHYTIN B, ... | | Authors: | Li, L, Nachtergaele, S.H.M, Seddon, A.M, Tereshko, V, Ponomarenko, N, Ismagilov, R.F, Accelerated Technologies Center for Gene to 3D Structure (ATCG3D) | | Deposit date: | 2008-05-09 | | Release date: | 2008-07-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Simple host-guest chemistry to modulate the process of concentration and crystallization of membrane proteins by detergent capture in a microfluidic device.
J.Am.Chem.Soc., 130, 2008
|
|
3DAN
 
 | | Crystal Structure of Allene oxide synthase | | Descriptor: | Cytochrome P450 74A2, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Li, L, Wang, X. | | Deposit date: | 2008-05-29 | | Release date: | 2008-09-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Modes of heme binding and substrate access for cytochrome P450 CYP74A revealed by crystal structures of allene oxide synthase.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
8FNZ
 
 | | Acetylated tau repeat 1 and 2 fragment (AcR1R2) | | Descriptor: | Microtubule-associated protein tau, acetylated repeat 1 and 2 fragment | | Authors: | Li, L, Nguyen, A.B, Mullapudi, V, Joachimiak, L. | | Deposit date: | 2022-12-29 | | Release date: | 2023-06-28 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.88 Å) | | Cite: | Disease-associated patterns of acetylation stabilize tau fibril formation.
Structure, 31, 2023
|
|
6KOO
 
 | | Mycobacterium tuberculosis initial transcription complex comprising sigma H and 5'-OH RNA of 7 nt | | Descriptor: | DNA (5'-D(*TP*GP*CP*AP*TP*CP*CP*GP*TP*GP*AP*GP*TP*CP*GP*AP*GP*GP*GP*TP*G)-3'), DNA (5'-D(*TP*TP*GP*TP*GP*GP*GP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*GP*CP*A)-3'), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Li, L, Zhang, Y. | | Deposit date: | 2019-08-12 | | Release date: | 2020-03-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | RNA extension drives a stepwise displacement of an initiation-factor structural module in initial transcription.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
3B9N
 
 | | Crystal structure of long-chain alkane monooxygenase (LadA) | | Descriptor: | Alkane monooxygenase | | Authors: | Li, L, Yang, W, Xu, F, Bartlam, M, Rao, Z. | | Deposit date: | 2007-11-06 | | Release date: | 2008-01-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of long-chain alkane monooxygenase (LadA) in complex with coenzyme FMN: unveiling the long-chain alkane hydroxylase
J.Mol.Biol., 376, 2008
|
|
3B9O
 
 | | long-chain alkane monooxygenase (LadA) in complex with coenzyme FMN | | Descriptor: | Alkane monooxygenase, FLAVIN MONONUCLEOTIDE | | Authors: | Li, L, Yang, W, Xu, F, Bartlam, M, Rao, Z. | | Deposit date: | 2007-11-06 | | Release date: | 2008-01-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of long-chain alkane monooxygenase (LadA) in complex with coenzyme FMN: unveiling the long-chain alkane hydroxylase
J.Mol.Biol., 376, 2008
|
|
3ZIU
 
 | | Crystal structure of Mycoplasma mobile Leucyl-tRNA Synthetase with Leu-AMS in the active site | | Descriptor: | 5'-O-(L-leucylsulfamoyl)adenosine, GLYCEROL, LEUCYL-TRNA SYNTHETASE | | Authors: | Li, L, Palencia, A, Lukk, T, Li, Z, Luthey-Schulten, Z.A, Cusack, S, Martinis, S.A, Boniecki, M.T. | | Deposit date: | 2013-01-10 | | Release date: | 2013-02-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Leucyl-tRNA Synthetase Editing Domain Functions as a Molecular Rheostat to Control Codon Ambiguity in Mycoplasma Pathogens.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
5DY3
 
 | | Crystal structure of Dbr2 | | Descriptor: | Artemisinic aldehyde Delta(11(13)) reductase, DI(HYDROXYETHYL)ETHER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Li, L, Chen, T.T, Xu, Y.C. | | Deposit date: | 2015-09-24 | | Release date: | 2016-10-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | The Crystal structure of Dbr2
to be published
|
|
5DXX
 
 | | Crystal structure of Dbr2 | | Descriptor: | Artemisinic aldehyde Delta(11(13)) reductase, FLAVIN MONONUCLEOTIDE | | Authors: | Li, L, Chen, T.T, Xu, Y.C. | | Deposit date: | 2015-09-24 | | Release date: | 2016-10-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The Crystal structure of Dbr2
to be published
|
|
5DXY
 
 | | Crystal structure of Dbr2 | | Descriptor: | Artemisinic aldehyde Delta(11(13)) reductase, FLAVIN MONONUCLEOTIDE, [[(2R,3S,4R,5R)-5-(5-aminocarbonyl-3,4-dihydro-2H-pyridin-1-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2R,3R,4R,5R)-5-(6-aminopurin-9-yl)-3-oxidanyl-4-phosphonooxy-oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Li, L, Chen, T.T, Xu, Y.C. | | Deposit date: | 2015-09-24 | | Release date: | 2016-10-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | The Crystal structure of Dbr2
to be published
|
|
5DY2
 
 | | Crystal structure of Dbr2 with mutation M27L | | Descriptor: | 1,6-DIHYDROXY NAPHTHALENE, Artemisinic aldehyde Delta(11(13)) reductase, FLAVIN MONONUCLEOTIDE | | Authors: | Li, L, Chen, T.T, Xu, Y.C. | | Deposit date: | 2015-09-24 | | Release date: | 2016-10-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | The Crystal structure of Dbr2
to be published
|
|
5NKM
 
 | | SMG8-SMG9 complex | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Li, L, Basquin, J, Conti, E. | | Deposit date: | 2017-03-31 | | Release date: | 2017-04-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.493 Å) | | Cite: | Structure of a SMG8-SMG9 complex identifies a G-domain heterodimer in the NMD effector proteins.
RNA, 23, 2017
|
|
5NKK
 
 | | SMG8-SMG9 complex GDP bound | | Descriptor: | 1,2-ETHANEDIOL, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Li, L, Basquin, J, Conti, E. | | Deposit date: | 2017-03-31 | | Release date: | 2017-04-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Structure of a SMG8-SMG9 complex identifies a G-domain heterodimer in the NMD effector proteins.
RNA, 23, 2017
|
|
2PQ6
 
 | |
1QVO
 
 | | STRUCTURES OF HLA-A*1101 IN COMPLEX WITH IMMUNODOMINANT NONAMER AND DECAMER HIV-1 EPITOPES CLEARLY REVEAL THE PRESENCE OF A MIDDLE ANCHOR RESIDUE | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-11 alpha chain, ... | | Authors: | Li, L, McNicholl, J.M, Bouvier, M. | | Deposit date: | 2003-08-28 | | Release date: | 2004-06-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structures of HLA-A*1101 complexed with immunodominant nonamer and decamer HIV-1 epitopes clearly reveal the presence of a middle, secondary anchor residue.
J.Immunol., 172, 2004
|
|
3DBM
 
 | | Crystal Structure of Allene oxide synthase | | Descriptor: | (9E,11E,13S)-13-hydroxyoctadeca-9,11-dienoic acid, Cytochrome P450 74A2, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Li, L, Wang, X. | | Deposit date: | 2008-06-01 | | Release date: | 2008-09-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Modes of heme binding and substrate access for cytochrome P450 CYP74A revealed by crystal structures of allene oxide synthase.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3DAM
 
 | | Crystal Structure of Allene oxide synthase | | Descriptor: | Cytochrome P450 74A2, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Li, L, Wang, X. | | Deposit date: | 2008-05-29 | | Release date: | 2008-09-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Modes of heme binding and substrate access for cytochrome P450 CYP74A revealed by crystal structures of allene oxide synthase.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
6P27
 
 | | Structure of a nested set of N-terminally extended MHC I-peptides provides novel insights into antigen processing and presentation | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, B-8 alpha chain, ... | | Authors: | Li, L, Batliwala, M, Bouvier, M. | | Deposit date: | 2019-05-21 | | Release date: | 2019-10-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.593 Å) | | Cite: | ERAP1 enzyme-mediated trimming and structural analyses of MHC I-bound precursor peptides yield novel insights into antigen processing and presentation.
J.Biol.Chem., 294, 2019
|
|
6P2C
 
 | | Structure of a nested set of N-terminally extended MHC I-peptides provides novel insights into antigen processing and presentation | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, B-8 alpha chain, ... | | Authors: | Li, L, Batliwala, M, Bouvier, M. | | Deposit date: | 2019-05-21 | | Release date: | 2019-10-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.396 Å) | | Cite: | ERAP1 enzyme-mediated trimming and structural analyses of MHC I-bound precursor peptides yield novel insights into antigen processing and presentation.
J.Biol.Chem., 294, 2019
|
|
6P2S
 
 | |
6P2F
 
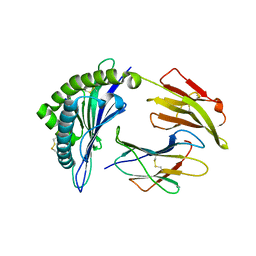 | | Structure of a nested set of N-terminally extended MHC I-peptides provides novel insights into antigen processing and presentation | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, B-8 alpha chain, ... | | Authors: | Li, L, Batliwala, M, Bouvier, M. | | Deposit date: | 2019-05-21 | | Release date: | 2019-10-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.482 Å) | | Cite: | ERAP1 enzyme-mediated trimming and structural analyses of MHC I-bound precursor peptides yield novel insights into antigen processing and presentation.
J.Biol.Chem., 294, 2019
|
|
6P23
 
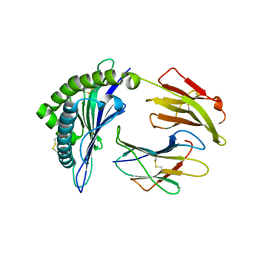 | | Structure of a nested set of N-terminally extended MHC I-peptides provide novel insights into antigen processing presentation | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, B-8 alpha chain, ... | | Authors: | Li, L, Batliwala, M, Bouvier, M. | | Deposit date: | 2019-05-20 | | Release date: | 2019-10-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.595 Å) | | Cite: | ERAP1 enzyme-mediated trimming and structural analyses of MHC I-bound precursor peptides yield novel insights into antigen processing and presentation.
J.Biol.Chem., 294, 2019
|
|
7XB0
 
 | | Crystal structure of Omicron BA.2 RBD complexed with hACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Li, L, Liao, H, Meng, Y, Li, W. | | Deposit date: | 2022-03-19 | | Release date: | 2022-07-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of human ACE2 higher binding affinity to currently circulating Omicron SARS-CoV-2 sub-variants BA.2 and BA.1.1.
Cell, 185, 2022
|
|
3C6D
 
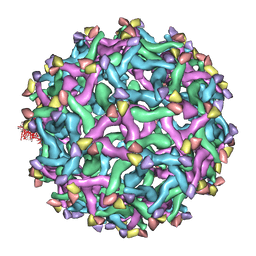 | | The pseudo-atomic structure of dengue immature virus | | Descriptor: | Polyprotein, prM | | Authors: | Li, L. | | Deposit date: | 2008-02-04 | | Release date: | 2008-04-08 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (12.5 Å) | | Cite: | The flavivirus precursor membrane-envelope protein complex: structure and maturation
Science, 319, 2008
|
|
3C5X
 
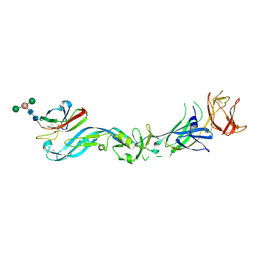 | | Crystal structure of the precursor membrane protein- envelope protein heterodimer from the dengue 2 virus at low pH | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope protein E, beta-D-mannopyranose-(1-3)-[beta-D-mannopyranose-(1-6)]alpha-D-altropyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Li, L. | | Deposit date: | 2008-02-01 | | Release date: | 2008-04-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The flavivirus precursor membrane-envelope protein complex: structure and maturation.
Science, 319, 2008
|
|
