3LET
 
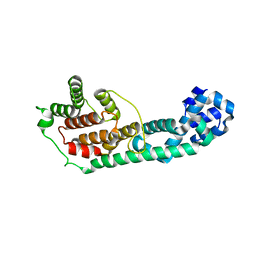 | | Crystal Structure of Fic domain containing AMPylator, VopS | | Descriptor: | Adenosine monophosphate-protein transferase vopS | | Authors: | Luong, P.H, Kinch, L.N, Brautigam, C.A, Grishin, N.V, Tomchick, D.R, Orth, K. | | Deposit date: | 2010-01-15 | | Release date: | 2010-04-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Structural and Kinetic Analysis of VopS with Fic Domain Supports a Direct Transfer Mechanism for AMPylation
To be Published
|
|
5DO7
 
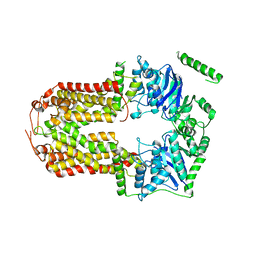 | | Crystal Structure of the Human Sterol Transporter ABCG5/ABCG8 | | Descriptor: | ATP-binding cassette sub-family G member 5, ATP-binding cassette sub-family G member 8 | | Authors: | Lee, J.-Y, Kinch, L.N, Borek, D.M, Urbatsch, I.L, Xie, X.-S, Grishin, N.V, Cohen, J.C, Otwinowski, Z, Hobbs, H.H, Rosenbaum, D.M. | | Deposit date: | 2015-09-10 | | Release date: | 2016-05-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.93 Å) | | Cite: | Crystal structure of the human sterol transporter ABCG5/ABCG8.
Nature, 533, 2016
|
|
5KEV
 
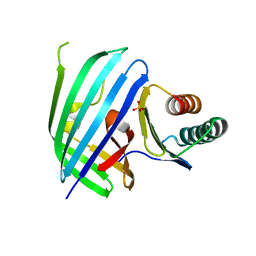 | |
5KEW
 
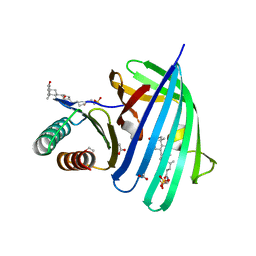 | | Vibrio parahaemolyticus VtrA/VtrC complex bound to the bile salt taurodeoxycholate | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, SULFATE ION, ... | | Authors: | Tomchick, D.R, Orth, K, Rivera-Cancel, G. | | Deposit date: | 2016-06-10 | | Release date: | 2016-07-20 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.103 Å) | | Cite: | Bile salt receptor complex activates a pathogenic type III secretion system.
Elife, 5, 2016
|
|
8DML
 
 | | Vibrio parahaemolyticus VtrA/VtrC complex bound to the bile salt chenodeoxycholate | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHENODEOXYCHOLIC ACID, ... | | Authors: | Tomchick, D.R, Orth, K, Zou, A.J. | | Deposit date: | 2022-07-08 | | Release date: | 2023-06-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Molecular determinants for differential activation of the bile acid receptor from the pathogen Vibrio parahaemolyticus.
J.Biol.Chem., 299, 2023
|
|
2LRU
 
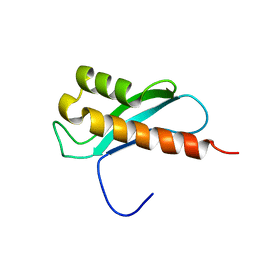 | | Solution Structure of the WNK1 Autoinhibitory Domain | | Descriptor: | Serine/threonine-protein kinase WNK1 | | Authors: | Moon, T.M, Correa, F, Gardner, K.H, Goldsmith, E.J. | | Deposit date: | 2012-04-13 | | Release date: | 2012-05-23 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the WNK1 Autoinhibitory Domain, a WNK-Specific PF2 Domain.
J.Mol.Biol., 425, 2013
|
|
5GZA
 
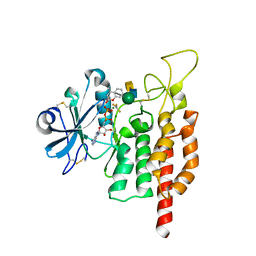 | | protein O-mannose kinase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-alpha-D-mannopyranose, 4-METHYL-2H-CHROMEN-2-ONE, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Xiao, J. | | Deposit date: | 2016-09-27 | | Release date: | 2016-12-07 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of protein O-mannose kinase reveals a unique active site architecture
Elife, 5, 2016
|
|
5TVM
 
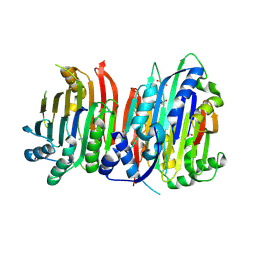 | | Crystal structure of Trypanosoma brucei AdoMetDC/prozyme heterodimer | | Descriptor: | 1,4-DIAMINOBUTANE, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, S-adenosylmethionine decarboxylase alpha chain, ... | | Authors: | Volkov, O.A, Chen, Z, Tomchick, D.R, Phillips, M.A. | | Deposit date: | 2016-11-09 | | Release date: | 2017-01-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.408 Å) | | Cite: | Relief of autoinhibition by conformational switch explains enzyme activation by a catalytically dead paralog.
Elife, 5, 2016
|
|
5TVO
 
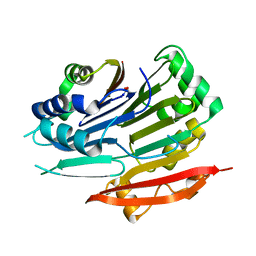 | | Crystal structure of Trypanosoma brucei AdoMetDC-delta26 monomer | | Descriptor: | PYRUVIC ACID, S-adenosylmethionine decarboxylase proenzyme, SODIUM ION | | Authors: | Volkov, O.A, Ariagno, C, Chen, Z, Tomchick, D.R, Phillips, M.A. | | Deposit date: | 2016-11-09 | | Release date: | 2016-12-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.481 Å) | | Cite: | Relief of autoinhibition by conformational switch explains enzyme activation by a catalytically dead paralog.
Elife, 5, 2016
|
|
4IIK
 
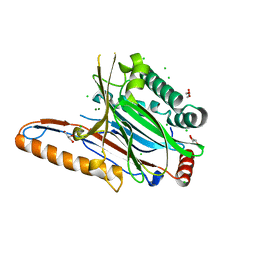 | | Legionella pneumophila effector | | Descriptor: | Adenosine monophosphate-protein hydrolase SidD, CHLORIDE ION, GLYCEROL, ... | | Authors: | Tascon, I, Chen, Y, Neunuebel, M.R, Rojas, A.L, Machner, M.P, Hierro, A. | | Deposit date: | 2012-12-20 | | Release date: | 2013-06-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Basis for Rab1 De-AMPylation by the Legionella pneumophila Effector SidD
Plos Pathog., 9, 2013
|
|
4IIP
 
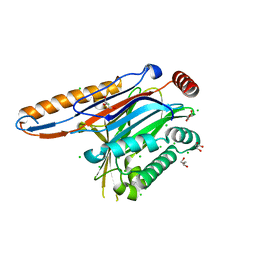 | | Legionella pneumophila effector | | Descriptor: | Adenosine monophosphate-protein hydrolase SidD, CHLORIDE ION, GLYCEROL | | Authors: | Tascon, I, Chen, Y, Neunuebel, M.R, Rojas, A.L, Machner, M.P, Hierro, A. | | Deposit date: | 2012-12-20 | | Release date: | 2013-06-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for Rab1 De-AMPylation by the Legionella pneumophila Effector SidD
Plos Pathog., 9, 2013
|
|
6EAC
 
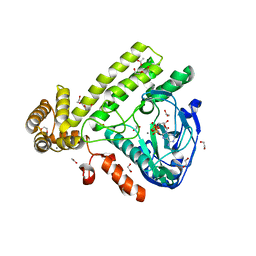 | | Pseudomonas syringae SelO | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Tomchick, D.R, Tagliabracci, V.S, Sreelatha, A. | | Deposit date: | 2018-08-02 | | Release date: | 2018-10-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.269 Å) | | Cite: | Protein AMPylation by an Evolutionarily Conserved Pseudokinase.
Cell, 175, 2018
|
|
7JLQ
 
 | | cryo-EM structure of human ATG9A in LMNG micelles | | Descriptor: | Autophagy-related protein 9A | | Authors: | Maeda, S, Otomo, T. | | Deposit date: | 2020-07-30 | | Release date: | 2020-10-28 | | Last modified: | 2020-12-16 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure, lipid scrambling activity and role in autophagosome formation of ATG9A.
Nat.Struct.Mol.Biol., 27, 2020
|
|
7JLO
 
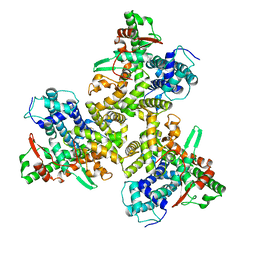 | | Cryo-EM structure of human ATG9A in amphipols | | Descriptor: | Autophagy-related protein 9A | | Authors: | Maeda, S, Otomo, T. | | Deposit date: | 2020-07-30 | | Release date: | 2020-10-28 | | Last modified: | 2020-12-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure, lipid scrambling activity and role in autophagosome formation of ATG9A.
Nat.Struct.Mol.Biol., 27, 2020
|
|
7JLP
 
 | | cryo-EM structure of human ATG9A in nanodiscs | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, Autophagy-related protein 9A | | Authors: | Maeda, S, Otomo, T. | | Deposit date: | 2020-07-30 | | Release date: | 2020-10-28 | | Last modified: | 2020-12-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure, lipid scrambling activity and role in autophagosome formation of ATG9A.
Nat.Struct.Mol.Biol., 27, 2020
|
|
3V5Y
 
 | | Structure of FBXL5 hemerythrin domain, P2(1) cell | | Descriptor: | F-box/LRR-repeat protein 5, MU-OXO-DIIRON | | Authors: | Tomchick, D.R, Bruick, R.K, Thompson, J.W, Brautigam, C.A. | | Deposit date: | 2011-12-17 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and Molecular Characterization of Iron-sensing Hemerythrin-like Domain within F-box and Leucine-rich Repeat Protein 5 (FBXL5).
J.Biol.Chem., 287, 2012
|
|
3V5Z
 
 | | Structure of FBXL5 hemerythrin domain, C2 cell, grown anaerobically | | Descriptor: | F-box/LRR-repeat protein 5, MU-OXO-DIIRON | | Authors: | Tomchick, D.R, Bruick, R.K, Thompson, J.W, Brautigam, C.A. | | Deposit date: | 2011-12-17 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1847 Å) | | Cite: | Structural and Molecular Characterization of Iron-sensing Hemerythrin-like Domain within F-box and Leucine-rich Repeat Protein 5 (FBXL5).
J.Biol.Chem., 287, 2012
|
|
3V5X
 
 | | Structure of FBXL5 hemerythrin domain, C2 cell | | Descriptor: | F-box/LRR-repeat protein 5, MU-OXO-DIIRON | | Authors: | Tomchick, D.R, Bruick, R.K, Thompson, J.W, Brautigam, C.A. | | Deposit date: | 2011-12-17 | | Release date: | 2012-01-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and Molecular Characterization of Iron-sensing Hemerythrin-like Domain within F-box and Leucine-rich Repeat Protein 5 (FBXL5).
J.Biol.Chem., 287, 2012
|
|
