1CVX
 
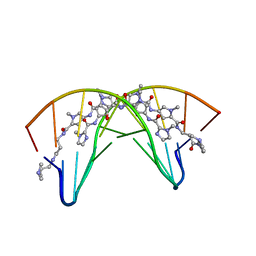 | | CRYSTAL STRUCTURE OF POLYAMIDE DIMER (IMPYHPPYBETADP)2 BOUND TO B-DNA DECAMER CCAGATCTGG | | Descriptor: | 5'-D(*CP*CP*AP*GP*AP*TP*CP*TP*GP*G)-3', HYDROXYPYRROLE-IMIDAZOLE-PYRROLE POLYAMIDE | | Authors: | Kielkopf, C.L, Bremer, R.E, White, S, Baird, E.E, Dervan, P.B, Rees, D.C. | | Deposit date: | 1999-08-24 | | Release date: | 2000-01-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structural effects of DNA sequence on T.A recognition by hydroxypyrrole/pyrrole pairs in the minor groove.
J.Mol.Biol., 295, 2000
|
|
1CVY
 
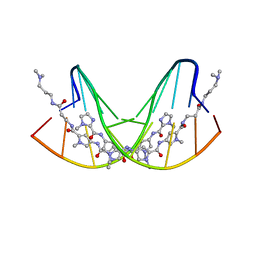 | | CRYSTAL STRUCTURE OF POLYAMIDE DIMER (IMPYPYPYBETADP)2 BOUND TO CCAGATCTGG | | Descriptor: | 5'-D(*CP*CP*AP*GP*AP*TP*CP*TP*GP*G)-3', IMIDAZOLE-PYRROLE POLYAMIDE | | Authors: | Kielkopf, C.L, Bremer, R.E, White, S, Baird, E.E, Dervan, P.B, Rees, D.C. | | Deposit date: | 1999-08-24 | | Release date: | 2000-01-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural effects of DNA sequence on T.A recognition by hydroxypyrrole/pyrrole pairs in the minor groove.
J.Mol.Biol., 295, 2000
|
|
1D8G
 
 | | ULTRAHIGH RESOLUTION CRYSTAL STRUCTURE OF B-DNA DECAMER D(CCAGTACTGG) | | Descriptor: | 5'-D(*CP*CP*AP*GP*TP*AP*CP*TP*GP*GP*)-3', CALCIUM ION | | Authors: | Kielkopf, C.L, Ding, S, Kuhn, P, Rees, D.C. | | Deposit date: | 1999-10-23 | | Release date: | 2000-02-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (0.74 Å) | | Cite: | Conformational flexibility of B-DNA at 0.74 A resolution: d(CCAGTACTGG)(2).
J.Mol.Biol., 296, 2000
|
|
1JG8
 
 | | Crystal Structure of Threonine Aldolase (Low-specificity) | | Descriptor: | CALCIUM ION, L-allo-threonine aldolase, SODIUM ION | | Authors: | Kielkopf, C.L, Bonanno, J, Ray, S, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2001-06-23 | | Release date: | 2001-07-04 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Low-specificity Threonine Aldolase, a Key Enzyme in Glycine Biosynthesis
To be published
|
|
1JMT
 
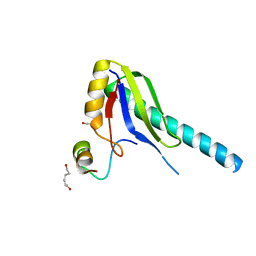 | | X-ray Structure of a Core U2AF65/U2AF35 Heterodimer | | Descriptor: | HEXANE-1,6-DIOL, SPLICING FACTOR U2AF 35 KDA SUBUNIT, SPLICING FACTOR U2AF 65 KDA SUBUNIT | | Authors: | Kielkopf, C.L, Rodionova, N.A, Green, M.R, Burley, S.K. | | Deposit date: | 2001-07-19 | | Release date: | 2001-09-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A novel peptide recognition mode revealed by the X-ray structure of a core U2AF35/U2AF65 heterodimer.
Cell(Cambridge,Mass.), 106, 2001
|
|
1LW4
 
 | |
1LW5
 
 | |
408D
 
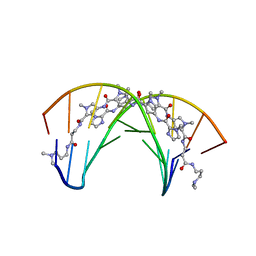 | | STRUCTURAL BASIS FOR RECOGNITION OF A-T AND T-A BASE PAIRS IN THE MINOR GROOVE OF B-DNA | | Descriptor: | DNA (5'-D(*CP*CP*AP*GP*TP*AP*CP*TP*GP*G)-3'), IMIDAZOLE-PYRROLE POLYAMIDE | | Authors: | Kielkopf, C.L, White, S, Szewczyk, J.W, Turner, J.M, Baird, E.E, Dervan, P.B, Rees, D.C. | | Deposit date: | 1998-06-24 | | Release date: | 1998-10-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A structural basis for recognition of A.T and T.A base pairs in the minor groove of B-DNA.
Science, 282, 1998
|
|
454D
 
 | | INTERCALATION AND MAJOR GROOVE RECOGNITION IN THE 1.2 A RESOLUTION CRYSTAL STRUCTURE OF RH[ME2TRIEN]PHI BOUND TO 5'-G(5IU)TGCAAC-3' | | Descriptor: | 5'-D(*GP*(5IU)P*TP*GP*CP*AP*AP*C)-3', DELTA-ALPHA-RH[2R,9R-DIAMINO-4,7-DIAZADECANE]9,10-PHENANTHRENEQUINONE DIIMINE | | Authors: | Kielkopf, C.L, Erkkila, K.E, Hudson, B.P, Barton, J.K, Rees, D.C. | | Deposit date: | 1999-03-03 | | Release date: | 2000-02-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure of a photoactive rhodium complex intercalated into DNA.
Nat.Struct.Biol., 7, 2000
|
|
365D
 
 | | STRUCTURAL BASIS FOR G C RECOGNITION IN THE DNA MINOR GROOVE | | Descriptor: | DNA (5'-D(*CP*CP*AP*GP*GP*(CBR)P*CP*TP*GP*G)-3'), IMIDAZOLE-PYRROLE POLYAMIDE | | Authors: | Kielkopf, C.L, Baird, E.E, Dervan, P.B, Rees, D.C. | | Deposit date: | 1997-12-17 | | Release date: | 1998-02-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for G.C recognition in the DNA minor groove.
Nat.Struct.Biol., 5, 1998
|
|
1M6S
 
 | | Crystal Structure Of Threonine Aldolase | | Descriptor: | CALCIUM ION, CHLORIDE ION, L-allo-threonine aldolase | | Authors: | Burley, S.K, Kielkopf, C.L. | | Deposit date: | 2002-07-17 | | Release date: | 2002-12-11 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray Structures of Threonine Aldolase Complexes: Structural Basis of Substrate Recognition
Biochemistry, 41, 2002
|
|
6XLW
 
 | | Crystal structure of U2AF65 bound to AdML splice site sequence | | Descriptor: | DI(HYDROXYETHYL)ETHER, DNA/RNA (5'-R(P*UP*UP*(UD)P*UP*U)-D(P*(BRU))-R(P*CP*C)-3'), GLYCEROL, ... | | Authors: | Maji, D, Jenkins, J.L, Kielkopf, C.L. | | Deposit date: | 2020-06-29 | | Release date: | 2020-10-07 | | Last modified: | 2022-03-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Representative cancer-associated U2AF2 mutations alter RNA interactions and splicing.
J.Biol.Chem., 295, 2020
|
|
6N3D
 
 | | Structure of HIV Tat-specific factor 1 U2AF Homology Motif (APO-State) | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Loerch, S, Jenkins, J.L, Kielkopf, C.L. | | Deposit date: | 2018-11-15 | | Release date: | 2019-01-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | The pre-mRNA splicing and transcription factor Tat-SF1 is a functional partner of the spliceosome SF3b1 subunit via a U2AF homology motif interface.
J. Biol. Chem., 294, 2019
|
|
1XCB
 
 | | X-ray Structure of a Rex-Family Repressor/NADH Complex from Thermus Aquaticus | | Descriptor: | CALCIUM ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Redox-sensing transcriptional repressor rex | | Authors: | Sickmier, E.A, Brekasis, D, Paranawithana, S, Bonanno, J.B, Burley, S.K, Paget, M.S, Kielkopf, C.L, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-09-01 | | Release date: | 2004-09-28 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | X-Ray Structure of a Rex-Family Repressor/NADH Complex: Insights into the Mechanism of Redox Sensing
Structure, 13, 2005
|
|
7S3A
 
 | |
7SN6
 
 | | U2AF65 UHM BOUND TO SF3B155 ULM5 | | Descriptor: | ISOPROPYL ALCOHOL, Splicing factor 3B subunit 1, Splicing factor U2AF 65 kDa subunit | | Authors: | Loerch, S, Jenkins, J.L, Kielkopf, C.L. | | Deposit date: | 2021-10-27 | | Release date: | 2022-07-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A UHM-ULM interface with unusual structural features contributes to U2AF2 and SF3B1 association for pre-mRNA splicing.
J.Biol.Chem., 298, 2022
|
|
2G4B
 
 | | Structure of U2AF65 variant with polyuridine tract | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, 5'-R(P*UP*UP*UP*UP*UP*UP*U)-3', Splicing factor U2AF 65 kDa subunit | | Authors: | Sickmier, E.A, Kielkopf, C.L. | | Deposit date: | 2006-02-21 | | Release date: | 2006-07-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of polypyrimidine tract recognition
by the essential splicing factor U2AF65.
Mol.Cell, 23, 2006
|
|
2HZC
 
 | |
6XLX
 
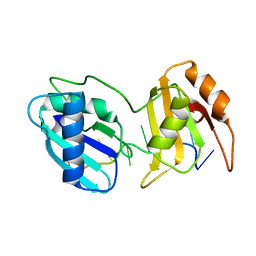 | |
6XLV
 
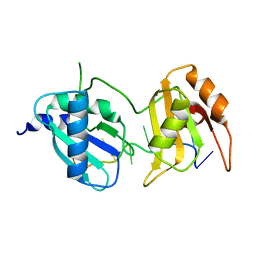 | |
2OKS
 
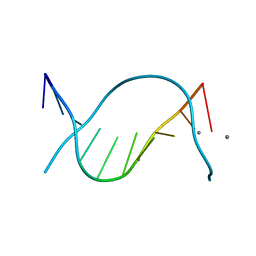 | | X-ray Structure of a DNA Repair Substrate Containing an Alkyl Interstrand Crosslink at 1.65 Resolution | | Descriptor: | 5'-D(*CP*CP*AP*AP*(C34)P*GP*TP*TP*GP*G)-3', CALCIUM ION | | Authors: | Swenson, M.C, Paranawithana, S.R, Miller, P.S, Kielkopf, C.L. | | Deposit date: | 2007-01-17 | | Release date: | 2007-02-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure of a DNA repair substrate containing an alkyl interstrand cross-link at 1.65 a resolution.
Biochemistry, 46, 2007
|
|
4FXX
 
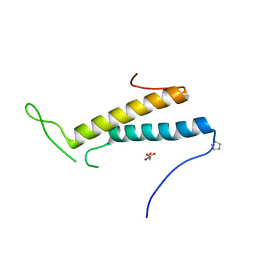 | | Structure of SF1 coiled-coil domain | | Descriptor: | IMIDAZOLE, MALONATE ION, Splicing factor 1 | | Authors: | Gupta, A, Bauer, W.J, Wang, W, Kielkopf, C.L. | | Deposit date: | 2012-07-03 | | Release date: | 2013-01-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4801 Å) | | Cite: | Structure of Phosphorylated SF1 Bound to U2AF(65) in an Essential Splicing Factor Complex.
Structure, 21, 2013
|
|
4FXW
 
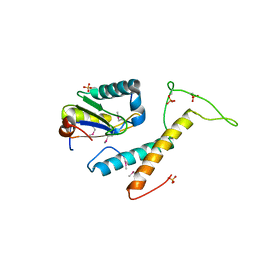 | | Structure of phosphorylated SF1 complex with U2AF65-UHM domain | | Descriptor: | SULFATE ION, Splicing factor 1, Splicing factor U2AF 65 kDa subunit | | Authors: | Wang, W, Bauer, W.J, Wedekind, J.E, Kielkopf, C.L. | | Deposit date: | 2012-07-03 | | Release date: | 2013-01-16 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structure of Phosphorylated SF1 Bound to U2AF(65) in an Essential Splicing Factor Complex.
Structure, 21, 2013
|
|
4TU9
 
 | | STRUCTURE OF U2AF65 VARIANT WITH BRU5G6 DNA | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, DNA (5'-D(*UP*UP*UP*UP*(BRU)P*DG*U)-3'), GLYCEROL, ... | | Authors: | Jenkins, J.L, McLaughlin, K.J, Agrawal, A.A, Kielkopf, C.L. | | Deposit date: | 2014-06-24 | | Release date: | 2014-11-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.992 Å) | | Cite: | Structure-guided U2AF65 variant improves recognition and splicing of a defective pre-mRNA.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4TU8
 
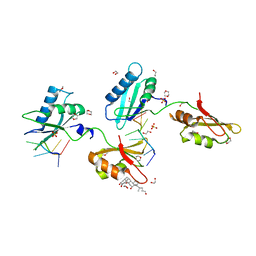 | | STRUCTURE OF U2AF65 VARIANT WITH BRU5A6 DNA | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIETHYLENE DIOXIDE, DNA (5'-D(*UP*UP*UP*UP*(BRU)P*DA*U)-3'), ... | | Authors: | MCLAUGHLIN, K.J, JENKINS, J.L, Agrawal, A.A, KIELKOPF, C.L. | | Deposit date: | 2014-06-24 | | Release date: | 2014-11-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.918 Å) | | Cite: | Structure-guided U2AF65 variant improves recognition and splicing of a defective pre-mRNA.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
