2X9P
 
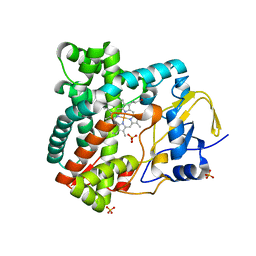 | | X-ray structure of the substrate-free cytochrome P450 PimD - a polyene macrolide antibiotic pimaricin epoxidase | | Descriptor: | PIMD PROTEIN, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Kells, P.M, Ouellet, H, Santos-Aberturas, J, Aparicio, J.F, Podust, L.M. | | Deposit date: | 2010-03-23 | | Release date: | 2010-08-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of Cytochrome P450 Pimd Suggests Epoxidation of the Polyene Macrolide Pimaricin Occurs Via a Hydroperoxoferric Intermediate.
Chem.Biol., 17, 2010
|
|
2XBK
 
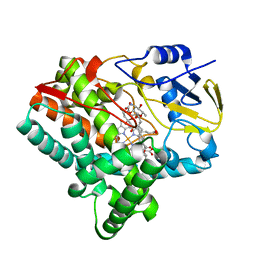 | | X-ray structure of the substrate-bound cytochrome P450 PimD - a polyene macrolide antibiotic pimaricin epoxidase | | Descriptor: | 4,5-DE-EPOXYPIMARICIN, PIMD PROTEIN, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Kells, P.M, Ouellet, H, Santos-Aberturas, J, Aparicio, J.F, Podust, L.M. | | Deposit date: | 2010-04-12 | | Release date: | 2010-08-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of Cytochrome P450 Pimd Suggests Epoxidation of the Polyene Macrolide Pimaricin Occurs Via a Hydroperoxoferric Intermediate.
Chem.Biol., 17, 2010
|
|
3ZSN
 
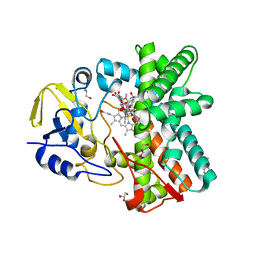 | | Structure of the mixed-function P450 MycG F286A mutant in complex with mycinamicin IV | | Descriptor: | BENZAMIDINE, GLYCEROL, MYCINAMICIN IV, ... | | Authors: | Li, S, Kells, P.M, Rutaganira, F.U, Anzai, Y, Kato, F, Sherman, D.H, Podust, L.M. | | Deposit date: | 2011-06-29 | | Release date: | 2012-05-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Substrate Recognition by the Multifunctional Cytochrome P450 Mycg in Mycinamicin Hydroxylation and Epoxidation Reactions.
J.Biol.Chem., 287, 2012
|
|
3ZBY
 
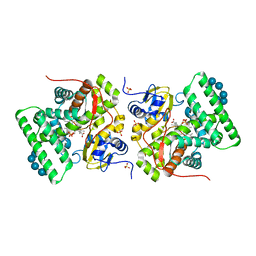 | | Ligand-free structure of CYP142 from Mycobacterium smegmatis | | Descriptor: | 1,2-ETHANEDIOL, Cycloheptakis-(1-4)-(alpha-D-glucopyranose), P450 HEME-THIOLATE PROTEIN, ... | | Authors: | Garcia-Fernandez, E, Frank, D.J, Galan, B, Kells, P.M, Podust, L.M, Garcia, J.L, Ortiz de Montellano, P.R. | | Deposit date: | 2012-11-13 | | Release date: | 2013-02-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | A Highly Conserved Mycobacterial Cholesterol Catabolic Pathway.
Environ.Microbiol., 15, 2013
|
|
4APY
 
 | | Ethylene glycol-bound form of P450 CYP125A3 from Mycobacterium smegmatis | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, P450 HEME-THIOLATE PROTEIN, ... | | Authors: | Frank, D.J, Garcia Fernandez, E, Kells, P.M, Garcia Lopez, J.L, Podust, L.M, Ortiz de Montellano, P.R. | | Deposit date: | 2012-04-09 | | Release date: | 2013-02-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Highly Conserved Mycobacterial Cholesterol Catabolic Pathway.
Environ.Microbiol., 15, 2013
|
|
4AW3
 
 | | Structure of the mixed-function P450 MycG F286V mutant in complex with mycinamicin V in P1 space group | | Descriptor: | GLYCEROL, MYCINAMICIN V, P-450-LIKE PROTEIN, ... | | Authors: | Li, S, Tietz, D.R, Rutaganira, F.U, Kells, P.M, Anzai, Y, Kato, F, Pochapsky, T.C, Sherman, D.H, Podust, L.M. | | Deposit date: | 2012-05-30 | | Release date: | 2012-09-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Substrate Recognition by the Multifunctional Cytochrome P450 Mycg in Mycinamicin Hydroxylation and Epoxidation Reactions.
J.Biol.Chem., 287, 2012
|
|
2WM4
 
 | | X-ray structure of Mycobacterium tuberculosis cytochrome P450 CYP124 in complex with phytanic acid | | Descriptor: | (3R,7S,11S)-3,7,11,15-tetramethylhexadecanoic acid, CALCIUM ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Johnston, J.B, Kells, P.M, Podust, L.M, Ortiz de Montellano, P.R. | | Deposit date: | 2009-06-30 | | Release date: | 2009-10-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Biochemical and structural characterization of CYP124: a methyl-branched lipid omega-hydroxylase from Mycobacterium tuberculosis.
Proc. Natl. Acad. Sci. U.S.A., 106, 2009
|
|
2X5W
 
 | | X-ray structure of Mycobacterium tuberculosis cytochrome P450 CYP125 in complex with substrate cholest-4-en-3-one | | Descriptor: | (8ALPHA,9BETA)-CHOLEST-4-EN-3-ONE, 1,2-ETHANEDIOL, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Ouellet, H, Guan, S, Johnston, J.B, Chow, E.D, Kells, P.M, Burlingame, A.L, Cox, J.S, Podust, L.M, Ortiz de Montellano, P.R. | | Deposit date: | 2010-02-10 | | Release date: | 2010-06-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Mycobacterium Tuberculosis Cyp125A1, a Steroid C27 Monooxygenase that Detoxifies Intracellularly Generated Cholest-4-En-3-One.
Mol.Microbiol., 77, 2010
|
|
2WM5
 
 | | X-ray structure of the substrate-free Mycobacterium tuberculosis cytochrome P450 CYP124 | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, PUTATIVE CYTOCHROME P450 124 | | Authors: | Johnston, J.B, Kells, P.M, Podust, L.M, Ortiz de Montellano, P.R. | | Deposit date: | 2009-06-30 | | Release date: | 2009-10-06 | | Last modified: | 2018-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Biochemical and structural characterization of CYP124: a methyl-branched lipid omega-hydroxylase from Mycobacterium tuberculosis.
Proc. Natl. Acad. Sci. U.S.A., 106, 2009
|
|
2X5L
 
 | | X-RAY STRUCTURE OF THE SUBSTRATE-FREE MYCOBACTERIUM TUBERCULOSIS CYTOCHROME P450 CYP125, ALTERNATIVE CRYSTAL FORM | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, PUTATIVE CYTOCHROME P450 125 | | Authors: | Ouellet, H, Kells, P.M, Ortiz de Montellano, P.R, Podust, L.M. | | Deposit date: | 2010-02-10 | | Release date: | 2010-11-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Reverse Type I Inhibitor of Mycobacteriumtuberculosis Cyp125A1.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
2Y5N
 
 | | Structure of the mixed-function P450 MycG in complex with mycinamicin V in P21 space group | | Descriptor: | GLYCEROL, MAGNESIUM ION, MYCINAMICIN V, ... | | Authors: | Li, S, Kells, P.M, Sherman, D.H, Podust, L.M. | | Deposit date: | 2011-01-15 | | Release date: | 2012-02-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Substrate Recognition by the Multifunctional Cytochrome P450 Mycg in Mycinamicin Hydroxylation and Epoxidation Reactions.
J.Biol.Chem., 287, 2012
|
|
2XC3
 
 | | X-ray structure of Mycobacterium tuberculosis cyp125 bound to the reverse type I inhibitor | | Descriptor: | NALPHA-[(TRANS-4-METHYLCYCLOHEXYL)CARBONYL]-N-PYRIDIN-4-YL-D-TRYPTOPHANAMIDE, PROTOPORPHYRIN IX CONTAINING FE, PUTATIVE CYTOCHROME P450 125 | | Authors: | Ouellet, H, Kells, P.M, Ortiz de Montellano, P.R, Podust, L.M. | | Deposit date: | 2010-04-16 | | Release date: | 2010-11-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Reverse Type I Inhibitor of Mycobacteriumtuberculosis Cyp125A1.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
2XN8
 
 | | X-RAY STRUCTURE OF THE SUBSTRATE-FREE MYCOBACTERIUM TUBERCULOSIS CYTOCHROME P450 CYP125 | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, PUTATIVE CYTOCHROME P450 125, SULFATE ION | | Authors: | Ouellet, H, Kells, P.M, Ortiz de Montellano, P.R, Podust, L.M. | | Deposit date: | 2010-07-30 | | Release date: | 2010-11-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Reverse Type I Inhibitor of Mycobacteriumtuberculosis Cyp125A1.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
2Y46
 
 | | Structure of the mixed-function P450 MycG in complex with mycinamicin IV in C 2 2 21 space group | | Descriptor: | BENZAMIDINE, GLYCEROL, MYCINAMICIN IV, ... | | Authors: | Li, S, Kells, P.M, Sherman, D.H, Podust, L.M. | | Deposit date: | 2011-01-05 | | Release date: | 2012-01-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Substrate Recognition by the Multifunctional Cytochrome P450 Mycg in Mycinamicin Hydroxylation and Epoxidation Reactions.
J.Biol.Chem., 287, 2012
|
|
2Y5Z
 
 | | Mixed-function P450 MycG in complex with mycinamicin III in C2221 space group | | Descriptor: | BENZAMIDINE, GLYCEROL, MYCINAMICIN III, ... | | Authors: | Li, S, Kells, P.M, Sherman, D.H, Podust, L.M. | | Deposit date: | 2011-01-19 | | Release date: | 2012-02-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Substrate Recognition by the Multifunctional Cytochrome P450 Mycg in Mycinamicin Hydroxylation and Epoxidation Reactions.
J.Biol.Chem., 287, 2012
|
|
2YCA
 
 | | Mixed-function P450 MycG in complex with mycinamicin III in P21212 space group | | Descriptor: | GLYCEROL, MYCINAMICIN III, P-450-LIKE PROTEIN, ... | | Authors: | Li, S, Kells, P.M, Sherman, D.H, Podust, L.M. | | Deposit date: | 2011-03-12 | | Release date: | 2012-03-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Substrate Recognition by the Multifunctional Cytochrome P450 Mycg in Mycinamicin Hydroxylation and Epoxidation Reactions.
J.Biol.Chem., 287, 2012
|
|
2Y98
 
 | | Structure of the mixed-function P450 MycG in complex with mycinamicin IV in P21212 space group | | Descriptor: | CHLORIDE ION, GLYCEROL, MYCINAMICIN IV, ... | | Authors: | Li, S, Kells, P.M, Sherman, D.H, Podust, L.M. | | Deposit date: | 2011-02-12 | | Release date: | 2012-02-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Substrate Recognition by the Multifunctional Cytochrome P450 Mycg in Mycinamicin Hydroxylation and Epoxidation Reactions.
J.Biol.Chem., 287, 2012
|
|
2YGX
 
 | | Structure of the mixed-function P450 MycG in P21 space group | | Descriptor: | GLYCEROL, P-450-LIKE PROTEIN, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Li, S, Kells, P.M, Rutaganira, F.U, Sherman, D.H, Podust, L.M. | | Deposit date: | 2011-04-22 | | Release date: | 2012-02-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Substrate Recognition by the Multifunctional Cytochrome P450 Mycg in Mycinamicin Hydroxylation and Epoxidation Reactions.
J.Biol.Chem., 287, 2012
|
|
2YOO
 
 | | Cholest-4-en-3-one bound structure of CYP142 from Mycobacterium smegmatis | | Descriptor: | (8ALPHA,9BETA)-CHOLEST-4-EN-3-ONE, MAGNESIUM ION, P450 HEME-THIOLATE PROTEIN, ... | | Authors: | Garcia-Fernandez, E, Frank, D.J, Galan, B, Kells, P.M, Podust, L.M, Garcia, J.L, Ortiz de Montellano, P.R. | | Deposit date: | 2012-10-25 | | Release date: | 2013-02-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | A Highly Conserved Mycobacterial Cholesterol Catabolic Pathway.
Environ.Microbiol., 15, 2013
|
|
5UHU
 
 | |
3ZK5
 
 | | PikC D50N mutant bound to the 10-DML analog with the 3-(N,N-dimethylamino)ethanoate anchoring group | | Descriptor: | (3R,4S,5S,7R,9E,11R,12R)-12-ethyl-3,5,7,11-tetramethyl-2,8-dioxooxacyclododec-9-en-4-yl N,N-dimethylglycinate, CYTOCHROME P450 HYDROXYLASE PIKC, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Podust, L.M. | | Deposit date: | 2013-01-21 | | Release date: | 2014-01-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Directing Group-Controlled Regioselectivity in an Enzymatic C-H Bond Oxygenation.
J.Am.Chem.Soc., 136, 2014
|
|
4B7S
 
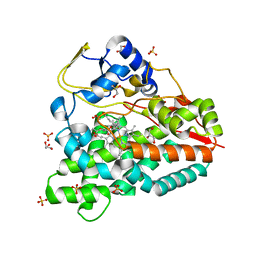 | |
4CCR
 
 | | Crystal structure of the thioredoxin reductase apoenzyme from Entamoeba histolytica in the absence of the NADP cofactor | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GOLD ION, THIOREDOXIN REDUCTASE | | Authors: | Parsonage, D, Kells, P.M, Hirata, K, Debnath, A, Poole, L.B, McKerrow, J.H, Reed, S.L, Podust, L.M. | | Deposit date: | 2013-10-25 | | Release date: | 2014-11-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | X-Ray Structures of Thioredoxin and Thioredoxin Reductase from Entamoeba Histolytica and Prevailing Hypothesis of the Mechanism of Auranofin Action.
J.Struct.Biol., 194, 2016
|
|
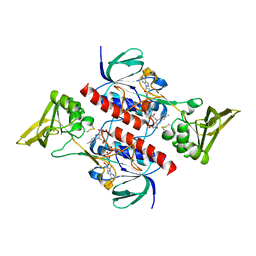
4CCQ
 
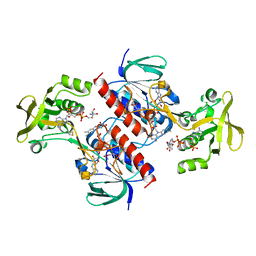 | | Crystal structure of the thioredoxin reductase from Entamoeba histolytica with NADP | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Parsonage, D, Kells, P.M, Hirata, K, Debnath, A, Poole, L.B, McKerrow, J.H, Reed, S.L, Podust, L.M. | | Deposit date: | 2013-10-24 | | Release date: | 2014-11-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | X-Ray Structures of Thioredoxin and Thioredoxin Reductase from Entamoeba Histolytica and Prevailing Hypothesis of the Mechanism of Auranofin Action.
J.Struct.Biol., 194, 2016
|
|
4CBQ
 
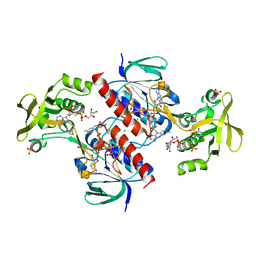 | | Crystal structure of the thioredoxin reductase from Entamoeba histolytica with auranofin Au(I) bound to Cys286 | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, GOLD ION, ... | | Authors: | Parsonage, D, Kells, P.M, Hirata, K, Debnath, A, Poole, L.B, McKerrow, J.H, Reed, S.L, Podust, L.M. | | Deposit date: | 2013-10-16 | | Release date: | 2014-11-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | X-Ray Structures of Thioredoxin and Thioredoxin Reductase from Entamoeba Histolytica and Prevailing Hypothesis of the Mechanism of Auranofin Action.
J.Struct.Biol., 194, 2016
|
|
