6LXK
 
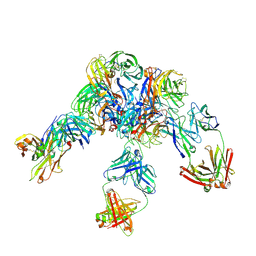 | | Crystal structure of Z2B3 D102R Fab in complex with influenza virus neuraminidase from A/Serbia/NS-601/2014 (H1N1) | | Descriptor: | CALCIUM ION, Heavy chain of Z2B3-D102R Fab, Light chain of Z2B3-D102R Fab, ... | | Authors: | Jiang, H, Peng, W, Qi, J, Chai, Y, Song, H, Shi, Y, Gao, G.F, Wu, Y. | | Deposit date: | 2020-02-11 | | Release date: | 2020-12-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.608 Å) | | Cite: | Structure-Based Modification of an Anti-neuraminidase Human Antibody Restores Protection Efficacy against the Drifted Influenza Virus.
Mbio, 11, 2020
|
|
6LXJ
 
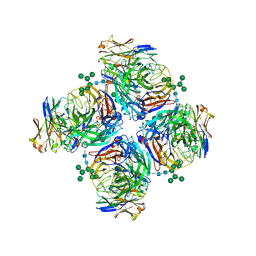 | | Crystal structure of human Z2B3 Fab in complex with influenza virus neuraminidase from A/Anhui/1/2013 (H7N9) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Heavy chain of Z2B3 Fab, ... | | Authors: | Jiang, H, Peng, W, Qi, J, Chai, Y, Song, H, Shi, Y, Gao, G.F, Wu, Y. | | Deposit date: | 2020-02-11 | | Release date: | 2020-12-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.903 Å) | | Cite: | Structure-Based Modification of an Anti-neuraminidase Human Antibody Restores Protection Efficacy against the Drifted Influenza Virus.
Mbio, 11, 2020
|
|
6LXI
 
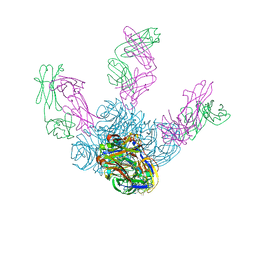 | | Crystal structure of Z2B3 Fab in complex with influenza virus neuraminidase from A/Brevig Mission/1/1918 (H1N1) | | Descriptor: | CALCIUM ION, Heavy chain of Z2B3 Fab, Light chain of Z2B3 Fab, ... | | Authors: | Jiang, H, Peng, W, Qi, J, Chai, Y, Song, H, Shi, Y, Gao, G.F, Wu, Y. | | Deposit date: | 2020-02-11 | | Release date: | 2020-12-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-Based Modification of an Anti-neuraminidase Human Antibody Restores Protection Efficacy against the Drifted Influenza Virus.
Mbio, 11, 2020
|
|
7WZC
 
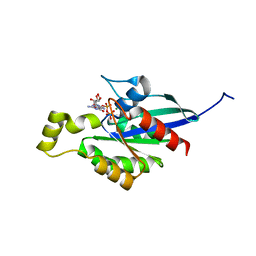 | | An open conformation Form2 of switch II for RhoA GDP-bound state | | Descriptor: | GLYCEROL, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Jiang, H, Luo, C. | | Deposit date: | 2022-02-17 | | Release date: | 2023-02-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.79944921 Å) | | Cite: | A RhoA structure with switch II flipped outward revealed the conformational dynamics of switch II region.
J.Struct.Biol., 215, 2023
|
|
7WZA
 
 | | An open conformation Form 1 of switch II for RhoA | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Transforming protein RhoA | | Authors: | Jiang, H, Luo, C. | | Deposit date: | 2022-02-17 | | Release date: | 2023-02-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.50028777 Å) | | Cite: | A RhoA structure with switch II flipped outward revealed the conformational dynamics of switch II region.
J.Struct.Biol., 215, 2023
|
|
5Y1Y
 
 | |
7CPD
 
 | | Crystal structure of T2R-TTL-(+)-6-Br-JP18 complex | | Descriptor: | (6R)-6-[(6-bromanyl-1H-indol-3-yl)methyl]-6,7,8,9-tetrahydrobenzo[7]annulen-5-one, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Jiang, H, Luo, C. | | Deposit date: | 2020-08-06 | | Release date: | 2021-08-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.506 Å) | | Cite: | Crystal structure of T2R-TTL-(+)-6-Br-JP18 complex
To Be Published
|
|
7CPQ
 
 | | crystal structure of T2R-TTL-(+)-6-Cl-JP18 complex | | Descriptor: | (6R)-6-[(6-chloranyl-1H-indol-3-yl)methyl]-6,7,8,9-tetrahydrobenzo[7]annulen-5-one, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Jiang, H, Luo, C. | | Deposit date: | 2020-08-07 | | Release date: | 2021-08-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.595 Å) | | Cite: | crystal structure of T2R-TTL-(+)-6-Cl-JP18 complex
To Be Published
|
|
7E9F
 
 | | Cryo-EM structure of the 2:1 Orc1 BAH domain in complex with nucleosome | | Descriptor: | DNA (147-mer), Histone H2A.2, Histone H2B.2, ... | | Authors: | Jiang, H, Yu, C, Liu, C.P, Han, X, Yu, Z, Xu, R.M. | | Deposit date: | 2021-03-04 | | Release date: | 2022-09-07 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Nucleosome binding relinquishes the association of the BAH domain of Orc1 with Sir1
To Be Published
|
|
7E9C
 
 | | Cryo-EM structure of the 1:1 Orc1 BAH domain in complex with nucleosome | | Descriptor: | DNA (147-mer), Histone H2A.2, Histone H2B.2, ... | | Authors: | Jiang, H, Yu, C, Liu, C.P, Han, X, Yu, Z, Xu, R.M. | | Deposit date: | 2021-03-04 | | Release date: | 2022-09-07 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Nucleosome binding relinquishes the association of the BAH domain of Orc1 with Sir1
To Be Published
|
|
5IZ6
 
 | | Protein-protein interaction | | Descriptor: | Adenomatous polyposis coli protein, DI(HYDROXYETHYL)ETHER, PHQ-ALA-GLY-GLU-ALA-LEU-TYR-GLU-NH2, ... | | Authors: | Zhao, Y, Jiang, H, Yang, X, Jiang, F, Song, K, Zhang, J. | | Deposit date: | 2016-03-25 | | Release date: | 2017-07-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Peptidomimetic inhibitors of APC-Asef interaction block colorectal cancer migration.
Nat. Chem. Biol., 13, 2017
|
|
8ZO0
 
 | |
5XM2
 
 | | Human N-terminal domain of FACT complex subunit SPT16 | | Descriptor: | DI(HYDROXYETHYL)ETHER, FACT complex subunit SPT16, GLYCEROL | | Authors: | Xu, S, Li, H, Dou, Y, Chen, Y, Jiang, H, Lu, D, Wang, M, Su, D. | | Deposit date: | 2017-05-12 | | Release date: | 2018-05-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.187 Å) | | Cite: | The structural basis of human Spt16 N-terminal domain interaction with histone (H3-H4)2tetramer.
Biochem.Biophys.Res.Commun., 508, 2019
|
|
9E6B
 
 | |
9E6C
 
 | | Octopus sensory receptor CRT1 in complex with H3C | | Descriptor: | 1-methyl-9H-pyrido[3,4-b]indole-3-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, Chemotactile receptor CRT1 | | Authors: | Jiang, H, Hibbs, R.E. | | Deposit date: | 2024-10-29 | | Release date: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Environmental microbiomes drive chemotactile sensation in octopus.
Cell, 2025
|
|
9E6D
 
 | |
5B6G
 
 | | Protein-protein interaction | | Descriptor: | Adenomatous polyposis coli protein, GLYCEROL, PHQ-ALA-GLY-GLU-ALA-XYC-TYR-GLU, ... | | Authors: | Zhao, Y, Jiang, H, Yang, X, Jiang, F, Song, K, Zhang, J. | | Deposit date: | 2016-05-27 | | Release date: | 2017-05-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Peptidomimetic inhibitors of APC-Asef interaction block colorectal cancer migration.
Nat. Chem. Biol., 13, 2017
|
|
6MS7
 
 | | Peroxisome proliferator-activated receptor gamma ligand binding domain in complex with a novel selective PPAR-gamma modulator VSP-77 | | Descriptor: | PGC1 LXXLL motif, Peroxisome proliferator-activated receptor gamma, {[(1S)-1-(4-chlorophenyl)octyl]oxy}acetic acid | | Authors: | Yi, W, Jiang, H, Zhou, X.E, Shi, J, Zhao, G, Zhang, X, Sun, Y, Suino-Powell, K, Li, J, Li, J, Melcher, K, Xu, H.E. | | Deposit date: | 2018-10-16 | | Release date: | 2019-10-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Identification and structural insight of an effective PPAR gamma modulator with improved therapeutic index for anti-diabetic drug discovery.
Chem Sci, 11, 2020
|
|
5IZ8
 
 | | Protein-protein interaction | | Descriptor: | ACE-ALA-GLY-GLU-ALA-LEU-ALA-ASP-NH2, Adenomatous polyposis coli protein, TRIETHYLENE GLYCOL | | Authors: | Zhao, Y, Jiang, H, Yang, X, Jiang, F, Song, K, Zhang, J. | | Deposit date: | 2016-03-25 | | Release date: | 2017-07-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Peptidomimetic inhibitors of APC-Asef interaction block colorectal cancer migration.
Nat. Chem. Biol., 13, 2017
|
|
5IZA
 
 | | Protein-protein interaction | | Descriptor: | ACE-GLY-GLY-GLU-ALA-LEU-ALA-TRP-NH2, Adenomatous polyposis coli protein | | Authors: | Zhao, Y, Jiang, H, Yang, X, Jiang, F, Song, K, Zhang, J. | | Deposit date: | 2016-03-25 | | Release date: | 2017-07-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Peptidomimetic inhibitors of APC-Asef interaction block colorectal cancer migration.
Nat. Chem. Biol., 13, 2017
|
|
5IZ9
 
 | | Protein-protein interaction | | Descriptor: | ACE-GLY-GLY-GLU-ALA-LEU-ALA-ASP-NH2, Adenomatous polyposis coli protein | | Authors: | Zhao, Y, Jiang, H, Yang, X, Jiang, F, Song, K, Zhang, J. | | Deposit date: | 2016-03-25 | | Release date: | 2017-07-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | Peptidomimetic inhibitors of APC-Asef interaction block colorectal cancer migration.
Nat. Chem. Biol., 13, 2017
|
|
8FRF
 
 | | Homodimeric designed loop protein RBL7_C2_3 | | Descriptor: | MALONATE ION, RBL7_C2_3 | | Authors: | Jude, K.M, Jiang, H, Baker, D, Garcia, K.C. | | Deposit date: | 2023-01-06 | | Release date: | 2024-04-17 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | De novo design of buttressed loops for sculpting protein functions.
Nat.Chem.Biol., 20, 2024
|
|
8FRE
 
 | | Designed loop protein RBL4 | | Descriptor: | 1,2-ETHANEDIOL, RBL4 | | Authors: | Jude, K.M, Jiang, H, Baker, D, Garcia, K.C. | | Deposit date: | 2023-01-06 | | Release date: | 2024-04-17 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | De novo design of buttressed loops for sculpting protein functions.
Nat.Chem.Biol., 20, 2024
|
|
7Y7I
 
 | | chicken KNL2 in complex with the CENP-A nucleosome | | Descriptor: | Chains: I, Chains: J, Histone H2A type 1-B/E, ... | | Authors: | Ariyoshi, M, Jiang, H, Makino, F, Fukagawa, T. | | Deposit date: | 2022-06-22 | | Release date: | 2022-07-13 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | The cryo-EM structure of the CENP-A nucleosome in complex with ggKNL2.
Biorxiv, 2022
|
|
3D04
 
 | | Crystal structure of (3R)-Hydroxyacyl-Acyl Carrier Protein Dehydratase (FabZ) from Helicobacter pylori in complex with sakuranetin | | Descriptor: | (2S)-5-hydroxy-2-(4-hydroxyphenyl)-7-methoxy-2,3-dihydro-4H-chromen-4-one, (3R)-hydroxymyristoyl-acyl carrier protein dehydratase, BENZAMIDINE, ... | | Authors: | Zhang, L, Kong, Y, Wu, D, Shen, X, Jiang, H. | | Deposit date: | 2008-05-01 | | Release date: | 2008-12-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Three flavonoids targeting the beta-hydroxyacyl-acyl carrier protein dehydratase from Helicobacter pylori: crystal structure characterization with enzymatic inhibition assay
Protein Sci., 17, 2008
|
|
