2VNN
 
 | | Human BACE-1 in complex with 7-ethyl-N-((1S,2R)-2-hydroxy-1-(phenylmethyl)-3-(((3-(trifluoromethyl)phenyl)methyl)amino)propyl)-1- methyl-3,4-dihydro-1H-(1,2,5)thiadiazepino(3,4,5-hi)indole-9- carboxamide 2,2-dioxide | | Descriptor: | BETA-SECRETASE 1, N-[(1S,2R)-1-benzyl-2-hydroxy-3-{[3-(trifluoromethyl)benzyl]amino}propyl]-7-ethyl-1-methyl-3,4-dihydro-1H-[1,2,5]thiadiazepino[3,4,5-hi]indole-9-carboxamide 2,2-dioxide | | Authors: | Charrier, N, Clarke, B, Cutler, L, Demont, E, Dingwall, C, Dunsdon, R, East, P, Hawkins, J, Howes, C, Hussain, I, Jeffrey, P, Maile, G, Matico, R, Mosley, J, Naylor, A, OBrien, A, Redshaw, S, Rowland, P, Soleil, V, Smith, K.J, Sweitzer, S, Theobald, P, Vesey, D, Walter, D.S, Wayne, G. | | Deposit date: | 2008-02-05 | | Release date: | 2008-05-20 | | Last modified: | 2019-05-15 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Second Generation of Hydroxyethylamine Bace-1 Inhibitors: Optimizing Potency and Oral Bioavailability.
J.Med.Chem., 51, 2008
|
|
2VNM
 
 | | Human BACE-1 in complex with 3-(1,1-dioxidotetrahydro-2H-1,2-thiazin- 2-yl)-5-(ethylamino)-N-((1S,2R)-2-hydroxy-1-(phenylmethyl)-3-(((3-(trifluoromethyl)phenyl)methyl)amino)propyl)benzamide | | Descriptor: | BETA-SECRETASE 1, N-[(1S,2R)-1-benzyl-2-hydroxy-3-{[3-(trifluoromethyl)benzyl]amino}propyl]-3-(1,1-dioxido-1,2-thiazinan-2-yl)-5-(ethylamino)benzamide | | Authors: | Charrier, N, Clarke, B, Cutler, L, Demont, E, Dingwall, C, Dunsdon, R, East, P, Hawkins, J, Howes, C, Hussain, I, Jeffrey, P, Maile, G, Matico, R, Mosley, J, Naylor, A, OBrien, A, Redshaw, S, Rowland, P, Soleil, V, Smith, K.J, Sweitzer, S, Theobald, P, Vesey, D, Walter, D.S, Wayne, G. | | Deposit date: | 2008-02-05 | | Release date: | 2008-05-20 | | Last modified: | 2019-05-15 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Second Generation of Hydroxyethylamine Bace-1 Inhibitors: Optimizing Potency and Oral Bioavailability.
J.Med.Chem., 51, 2008
|
|
3H4M
 
 | | AAA ATPase domain of the proteasome- activating nucleotidase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Proteasome-activating nucleotidase | | Authors: | Jeffrey, P, Zhang, F, Hu, M, Tian, G, Zhang, P, Finley, D, Shi, Y. | | Deposit date: | 2009-04-20 | | Release date: | 2009-06-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.106 Å) | | Cite: | Structural Insights into the Regulatory Particle of the Proteasome from Methanocaldococcus jannaschii.
Mol.Cell, 34, 2009
|
|
7Z8O
 
 | | Crystal structure of SARS-CoV-2 S RBD in complex with a stapled peptide | | Descriptor: | 2,4,6-tris(chloromethyl)-1,3,5-triazine, GLYCEROL, Spike protein S1, ... | | Authors: | Brear, P, Chen, L, Gaynor, K, Harman, M, Dods, R, Hyvonen, M. | | Deposit date: | 2022-03-18 | | Release date: | 2023-06-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (0.96 Å) | | Cite: | Multivalent bicyclic peptides are an effective antiviral modality that can potently inhibit SARS-CoV-2.
Nat Commun, 14, 2023
|
|
8AAA
 
 | | Crystal structure of SARS-CoV-2 S RBD in complex with a stapled peptide | | Descriptor: | 1,1',1''-(1,3,5-triazinane-1,3,5-triyl)tripropan-1-one, Spike protein S1, Stapled peptide | | Authors: | Brear, P, Chen, L, Gaynor, K, Harman, M, Dods, R, Hyvonen, M. | | Deposit date: | 2022-06-30 | | Release date: | 2023-06-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Multivalent bicyclic peptides are an effective antiviral modality that can potently inhibit SARS-CoV-2.
Nat Commun, 14, 2023
|
|
6Z7B
 
 | | Variant Surface Glycoprotein VSGsur bound to suramin | | Descriptor: | 8,8'-[CARBONYLBIS[IMINO-3,1-PHENYLENECARBONYLIMINO(4-METHYL-3,1-PHENYLENE)CARBONYLIMINO]]BIS-1,3,5-NAPHTHALENETRISULFON IC ACID, Variant surface glycoprotein Sur, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zeelen, J.P, Straaten van, M, Stebbins, C.E, Jeffrey, P. | | Deposit date: | 2020-05-30 | | Release date: | 2021-03-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structure of trypanosome coat protein VSGsur and function in suramin resistance.
Nat Microbiol, 6, 2021
|
|
4C66
 
 | | Discovery of Epigenetic Regulator I-BET762: Lead Optimization to Afford a Clinical Candidate Inhibitor of the BET Bromodomains | | Descriptor: | 4-(2-chlorophenyl)-2-ethyl-9-methyl-6,8-dihydrothieno[3,2-f][1,2,4]triazolo[4,3-a][1,4]diazepin-10-ium, BROMODOMAIN-CONTAINING PROTEIN 4 | | Authors: | Chung, C, Mirguet, O. | | Deposit date: | 2013-09-17 | | Release date: | 2013-10-02 | | Last modified: | 2013-10-30 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Discovery of Epigenetic Regulator I-Bet762: Lead Optimization to Afford a Clinical Candidate Inhibitor of the Bet Bromodomains.
J.Med.Chem., 56, 2013
|
|
4C67
 
 | | Discovery of Epigenetic Regulator I-BET762: Lead Optimization to Afford a Clinical Candidate Inhibitor of the BET Bromodomains | | Descriptor: | 1,2-ETHANEDIOL, 13-methyl-7-phenyl-3-thia-1,8,11,12-tetraazatricyclo trideca-2(6),4,7,10,12-pentaene, BROMODOMAIN-CONTAINING PROTEIN 4 | | Authors: | Chung, C, Mirguet, O. | | Deposit date: | 2013-09-17 | | Release date: | 2013-10-02 | | Last modified: | 2013-10-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Discovery of Epigenetic Regulator I-Bet762: Lead Optimization to Afford a Clinical Candidate Inhibitor of the Bet Bromodomains.
J.Med.Chem., 56, 2013
|
|
1TSR
 
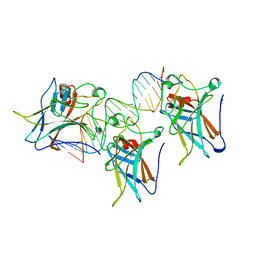 | | P53 CORE DOMAIN IN COMPLEX WITH DNA | | Descriptor: | DNA (5'-D(*AP*TP*AP*AP*TP*TP*GP*GP*GP*CP*AP*AP*GP*TP*CP*TP*A P*GP*GP*AP*A)-3'), DNA (5'-D(*TP*TP*TP*CP*CP*TP*AP*GP*AP*CP*TP*TP*GP*CP*CP*CP*A P*AP*TP*TP*A)-3'), PROTEIN (P53 TUMOR SUPPRESSOR), ... | | Authors: | Cho, Y, Gorina, S, Jeffrey, P, Pavletich, N. | | Deposit date: | 1995-07-28 | | Release date: | 1996-01-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a p53 tumor suppressor-DNA complex: understanding tumorigenic mutations.
Science, 265, 1994
|
|
3QP4
 
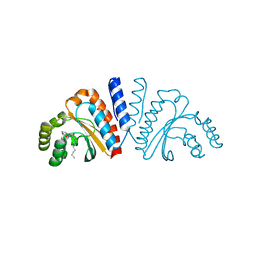 | | Crystal structure of CviR ligand-binding domain bound to C10-HSL | | Descriptor: | CviR transcriptional regulator, N-[(3S)-2-oxotetrahydrofuran-3-yl]decanamide | | Authors: | Chen, G, Swem, L, Swem, D, Stauff, D, O'Loughlin, C, Jeffrey, P, Bassler, B, Hughson, F. | | Deposit date: | 2011-02-11 | | Release date: | 2011-03-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A strategy for antagonizing quorum sensing.
Mol.Cell, 42, 2011
|
|
3QP2
 
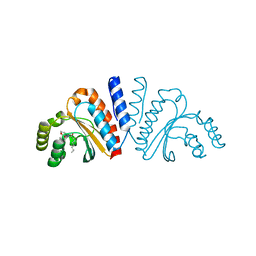 | | Crystal structure of CviR ligand-binding domain bound to C8-HSL | | Descriptor: | CviR transcriptional regulator, N-(2-OXOTETRAHYDROFURAN-3-YL)OCTANAMIDE | | Authors: | Chen, G, Swem, L, Swem, D, Stauff, D, O'Loughlin, C, Jeffrey, P, Bassler, B, Hughson, F. | | Deposit date: | 2011-02-11 | | Release date: | 2011-03-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.638 Å) | | Cite: | A strategy for antagonizing quorum sensing.
Mol.Cell, 42, 2011
|
|
3QP8
 
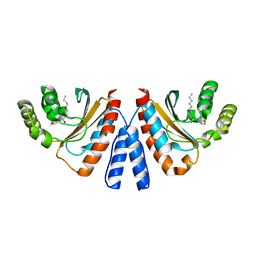 | | Crystal structure of CviR (Chromobacterium violaceum 12472) ligand-binding domain bound to C10-HSL | | Descriptor: | CviR transcriptional regulator, N-[(3S)-2-oxotetrahydrofuran-3-yl]decanamide | | Authors: | Chen, G, Swem, L, Swem, D, Stauff, D, O'Loughlin, C, Jeffrey, P, Bassler, B, Hughson, F. | | Deposit date: | 2011-02-11 | | Release date: | 2011-03-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A strategy for antagonizing quorum sensing.
Mol.Cell, 42, 2011
|
|
3QP5
 
 | | Crystal structure of CviR bound to antagonist chlorolactone (CL) | | Descriptor: | 4-(4-chlorophenoxy)-N-[(3S)-2-oxotetrahydrofuran-3-yl]butanamide, CviR transcriptional regulator | | Authors: | Chen, G, Swem, L, Swem, D, Stauff, D, O'Loughlin, C, Jeffrey, P, Bassler, B, Hughson, F. | | Deposit date: | 2011-02-11 | | Release date: | 2011-03-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.249 Å) | | Cite: | A strategy for antagonizing quorum sensing.
Mol.Cell, 42, 2011
|
|
3QP6
 
 | | Crystal structure of CviR (Chromobacterium violaceum 12472) bound to C6-HSL | | Descriptor: | CviR transcriptional regulator, N-[(3S)-2-oxotetrahydrofuran-3-yl]hexanamide | | Authors: | Chen, G, Swem, L, Swem, D, Stauff, D, O'Loughlin, C, Jeffrey, P, Bassler, B, Hughson, F. | | Deposit date: | 2011-02-11 | | Release date: | 2011-03-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A strategy for antagonizing quorum sensing.
Mol.Cell, 42, 2011
|
|
3QP1
 
 | | Crystal structure of CviR ligand-binding domain bound to the native ligand C6-HSL | | Descriptor: | CviR transcriptional regulator, N-[(3S)-2-oxotetrahydrofuran-3-yl]hexanamide | | Authors: | Chen, G, Swem, L, Swem, D, Stauff, D, O'Loughlin, C, Jeffrey, P, Bassler, B, Hughson, F. | | Deposit date: | 2011-02-11 | | Release date: | 2011-03-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A strategy for antagonizing quorum sensing.
Mol.Cell, 42, 2011
|
|
3ZYU
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with I-BET151(GSK1210151A) | | Descriptor: | 1,2-ETHANEDIOL, 7-(3,5-DIMETHYL-1,2-OXAZOL-4-YL)-8-METHOXY-1-[(1R)-1-(PYRIDIN-2-YL)ETHYL]-1H,2H,3H-IMIDAZO[4,5-C]QUINOLIN-2-ONE, BROMODOMAIN-CONTAINING PROTEIN 4 | | Authors: | Chung, C.W, Mirguet, O. | | Deposit date: | 2011-08-25 | | Release date: | 2011-11-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Inhibition of Bet Recruitment to Chromatin as an Effective Treatment for Mll-Fusion Leukaemia.
Nature, 478, 2011
|
|
4G0F
 
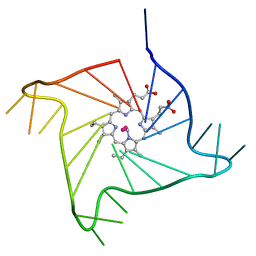 | | Crystal structure of the complex of a human telomeric repeat G-quadruplex and N-methyl mesoporphyrin IX (P6) | | Descriptor: | DNA (5'-D(*AP*GP*GP*GP*TP*TP*AP*GP*GP*GP*TP*TP*AP*GP*GP*GP*TP*TP*AP*GP*GP*G)-3'), N-METHYLMESOPORPHYRIN, POTASSIUM ION | | Authors: | Nicoludis, J.M, Miller, S.T, Jeffrey, P, Lawton, T.J, Rosenzweig, A.C, Yatsunyk, L.A. | | Deposit date: | 2012-07-09 | | Release date: | 2012-12-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Optimized End-Stacking Provides Specificity of N-Methyl Mesoporphyrin IX for Human Telomeric G-Quadruplex DNA.
J.Am.Chem.Soc., 134, 2012
|
|
4FXM
 
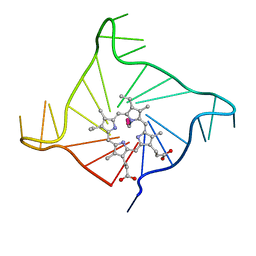 | | Crystal structure of the complex of a human telomeric repeat G-quadruplex and N-methyl mesoporphyrin IX (P21212) | | Descriptor: | DNA (5'-D(*AP*GP*GP*GP*TP*TP*AP*GP*GP*GP*TP*TP*AP*GP*GP*GP*TP*TP*AP*GP*GP*G)-3'), N-METHYLMESOPORPHYRIN, POTASSIUM ION | | Authors: | Nicoludis, J.M, Miller, S.T, Jeffrey, P, Lawton, T.J, Rosenzweig, A.C, Yatsunyk, L.A. | | Deposit date: | 2012-07-03 | | Release date: | 2012-12-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.651 Å) | | Cite: | Optimized End-Stacking Provides Specificity of N-Methyl Mesoporphyrin IX for Human Telomeric G-Quadruplex DNA.
J.Am.Chem.Soc., 134, 2012
|
|
6MYW
 
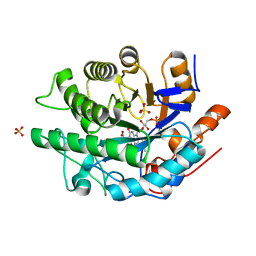 | | Gluconobacter Ene-Reductase (GluER) mutant - T36A | | Descriptor: | ACETATE ION, FLAVIN MONONUCLEOTIDE, GLYCEROL, ... | | Authors: | Garfinkle, S.E, Jeffrey, P, Hyster, T.K. | | Deposit date: | 2018-11-02 | | Release date: | 2019-06-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.157 Å) | | Cite: | Photoexcitation of flavoenzymes enables a stereoselective radical cyclization.
Science, 364, 2019
|
|
6O08
 
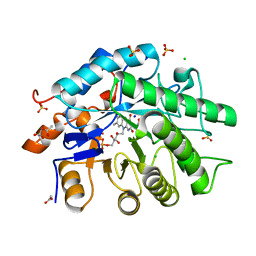 | | Gluconobacter Ene-Reductase (GluER) | | Descriptor: | ACETATE ION, CHLORIDE ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Garfinkle, S.E, Jeffrey, P, Hyster, T.K. | | Deposit date: | 2019-02-15 | | Release date: | 2019-06-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | Photoexcitation of flavoenzymes enables a stereoselective radical cyclization.
Science, 364, 2019
|
|
7TNB
 
 | | Caulobacter segnis arene reductase (CSAR) - WT | | Descriptor: | ACETATE ION, FLAVIN MONONUCLEOTIDE, NADH:flavin oxidoreductase/NADH oxidase, ... | | Authors: | Garfinkle, S.E, Jeffrey, P, Hyster, T.K. | | Deposit date: | 2022-01-20 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | An asymmetric sp 3 -sp 3 cross-electrophile coupling using 'ene'-reductases.
Nature, 610, 2022
|
|
