1IOP
 
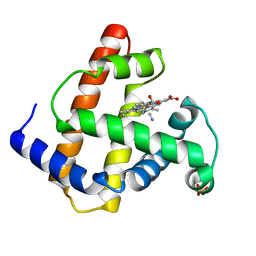 | | INCORPORATION OF A HEMIN WITH THE SHORTEST ACID SIDE-CHAINS INTO MYOGLOBIN | | Descriptor: | 6,7-DICARBOXYL-1,2,3,4,5,8-HEXAMETHYLHEMIN, CYANIDE ION, MYOGLOBIN, ... | | Authors: | Igarashi, N, Neya, S, Funasaki, N, Tanaka, N. | | Deposit date: | 1997-12-12 | | Release date: | 1998-04-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and function of 6,7-dicarboxyheme-substituted myoglobin
Biochemistry, 37, 1998
|
|
1FGJ
 
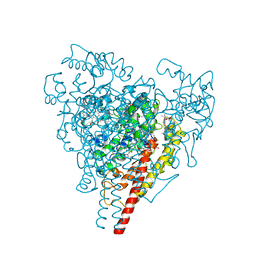 | | X-RAY STRUCTURE OF HYDROXYLAMINE OXIDOREDUCTASE | | Descriptor: | HEME C, HYDROXYLAMINE OXIDOREDUCTASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Tanaka, N, Igarashi, N, Moriyama, H. | | Deposit date: | 1997-03-03 | | Release date: | 1998-03-04 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The 2.8 A structure of hydroxylamine oxidoreductase from a nitrifying chemoautotrophic bacterium, Nitrosomonas europaea.
Nat.Struct.Biol., 4, 1997
|
|
1JWG
 
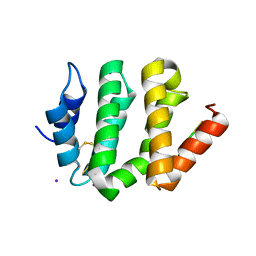 | | VHS Domain of human GGA1 complexed with cation-independent M6PR C-terminal Peptide | | Descriptor: | ADP-ribosylation factor binding protein GGA1, Cation-independent mannose-6-phosphate receptor, IODIDE ION | | Authors: | Shiba, T, Takatsu, H, Nogi, T, Matsugaki, N, Kawasaki, M, Igarashi, N, Suzuki, M, Kato, R, Earnest, T, Nakayama, K, Wakatsuki, S. | | Deposit date: | 2001-09-04 | | Release date: | 2002-03-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for recognition of acidic-cluster dileucine sequence by GGA1.
Nature, 415, 2002
|
|
1JWF
 
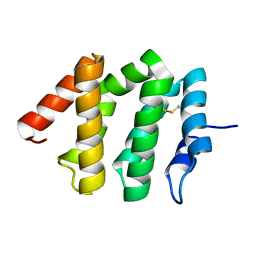 | | Crystal Structure of human GGA1 VHS domain. | | Descriptor: | ADP-ribosylation factor binding protein GGA1 | | Authors: | Shiba, T, Takatsu, H, Nogi, T, Matsugaki, N, Kawasaki, M, Igarashi, N, Suzuki, M, Kato, R, Earnest, T, Nakayama, K, Wakatsuki, S. | | Deposit date: | 2001-09-04 | | Release date: | 2002-03-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for recognition of acidic-cluster dileucine sequence by GGA1.
Nature, 415, 2002
|
|
1IU1
 
 | | Crystal structure of human gamma1-adaptin ear domain | | Descriptor: | gamma1-adaptin | | Authors: | Nogi, T, Shiba, Y, Kawasaki, M, Shiba, T, Matsugaki, N, Igarashi, N, Suzuki, M, Kato, R, Takatsu, H, Nakayama, K, Wakatsuki, S. | | Deposit date: | 2002-02-19 | | Release date: | 2002-07-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the accessory protein recruitment by the gamma-adaptin ear domain.
Nat.Struct.Biol., 9, 2002
|
|
1ITK
 
 | | Crystal structure of catalase-peroxidase from Haloarcula marismortui | | Descriptor: | CHLORIDE ION, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION, ... | | Authors: | Yamada, Y, Fujiwara, T, Sato, T, Igarashi, N, Tanaka, N. | | Deposit date: | 2002-01-18 | | Release date: | 2002-08-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The 2.0 A crystal structure of catalase-peroxidase from Haloarcula marismortui.
Nat.Struct.Biol., 9, 2002
|
|
1J2J
 
 | | Crystal structure of GGA1 GAT N-terminal region in complex with ARF1 GTP form | | Descriptor: | ADP-ribosylation factor 1, ADP-ribosylation factor binding protein GGA1, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Shiba, T, Kawasaki, M, Takatsu, H, Nogi, T, Matsugaki, N, Igarashi, N, Suzuki, M, Kato, R, Nakayama, K, Wakatsuki, S. | | Deposit date: | 2003-01-05 | | Release date: | 2003-05-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular mechanism of membrane recruitment of GGA by ARF in lysosomal protein transport
NAT.STRUCT.BIOL., 10, 2003
|
|
1O3Y
 
 | | Crystal structure of mouse ARF1 (delta17-Q71L), GTP form | | Descriptor: | ADP-ribosylation factor 1, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION | | Authors: | Shiba, T, Kawasaki, M, Takatsu, H, Nogi, T, Matsugaki, N, Igarashi, N, Suzuki, M, Kato, R, Nakayama, K, Wakatsuki, S. | | Deposit date: | 2003-05-08 | | Release date: | 2003-05-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Molecular mechanism of membrane recruitment of GGA by ARF in lysosomal protein transport
Nat.Struct.Biol., 10, 2003
|
|
1O3X
 
 | | Crystal structure of human GGA1 GAT domain | | Descriptor: | ADP-ribosylation factor binding protein GGA1 | | Authors: | Shiba, T, Kawasaki, M, Takatsu, H, Nogi, T, Matsugaki, N, Igarashi, N, Suzuki, M, Kato, R, Nakayama, K, Wakatsuki, S. | | Deposit date: | 2003-05-08 | | Release date: | 2003-05-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular Mechanism of Membrane Recruitment of Gga by Arf in Lysosomal Protein Transport
Nat.Struct.Biol., 10, 2003
|
|
2A6E
 
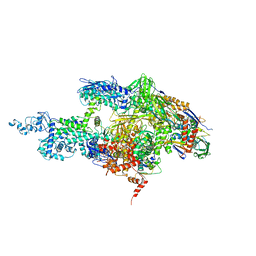 | | Crystal structure of the T. Thermophilus RNA polymerase holoenzyme | | Descriptor: | DNA-directed RNA polymerase alpha chain, DNA-directed RNA polymerase beta chain, DNA-directed RNA polymerase beta' chain, ... | | Authors: | Artsimovitch, I, Vassylyeva, M.N, Svetlov, D, Svetlov, V, Perederina, A, Igarashi, N, Matsugaki, N, Wakatsuki, S, Tahirov, T.H, Vassylyev, D.G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-02 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Allosteric modulation of the RNA polymerase catalytic reaction is an essential component of transcription control by rifamycins.
Cell(Cambridge,Mass.), 122, 2005
|
|
2A6H
 
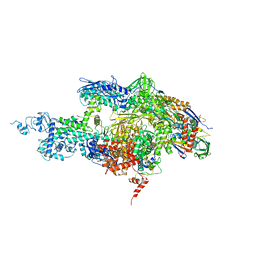 | | Crystal structure of the T. thermophilus RNA polymerase holoenzyme in complex with antibiotic sterptolydigin | | Descriptor: | DNA-directed RNA polymerase alpha chain, DNA-directed RNA polymerase beta chain, DNA-directed RNA polymerase beta' chain, ... | | Authors: | Temiakov, D, Zenkin, N, Vassylyeva, M.N, Perederina, A, Tahirov, T.H, Savkina, M, Zorov, S, Nikiforov, V, Igarashi, N, Matsugaki, N, Wakatsuki, S, Severinov, K, Vassylyev, D.G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-02 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of transcription inhibition by antibiotic streptolydigin.
Mol.Cell, 19, 2005
|
|
1WR6
 
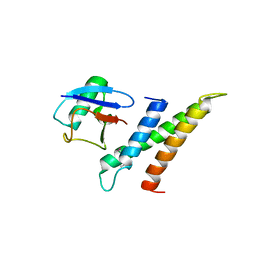 | | Crystal structure of GGA3 GAT domain in complex with ubiquitin | | Descriptor: | ADP-ribosylation factor binding protein GGA3, ubiquitin | | Authors: | Kawasaki, M, Shiba, T, Shiba, Y, Yamaguchi, Y, Matsugaki, N, Igarashi, N, Suzuki, M, Kato, R, Kato, K, Nakayama, K, Wakatsuki, S. | | Deposit date: | 2004-10-12 | | Release date: | 2005-06-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular mechanism of ubiquitin recognition by GGA3 GAT domain.
Genes Cells, 10, 2005
|
|
2A69
 
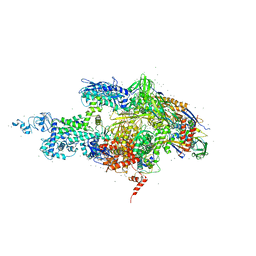 | | Crystal structure of the T. Thermophilus RNA polymerase holoenzyme in complex with antibiotic rifapentin | | Descriptor: | DNA-directed RNA polymerase alpha chain, DNA-directed RNA polymerase beta chain, DNA-directed RNA polymerase beta' chain, ... | | Authors: | Artsimovitch, I, Vassylyeva, M.N, Svetlov, D, Svetlov, V, Perederina, A, Igarashi, N, Matsugaki, N, Wakatsuki, S, Tahirov, T.H, Vassylyev, D.G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-02 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Allosteric modulation of the RNA polymerase catalytic reaction is an essential component of transcription control by rifamycins.
Cell(Cambridge,Mass.), 122, 2005
|
|
2A68
 
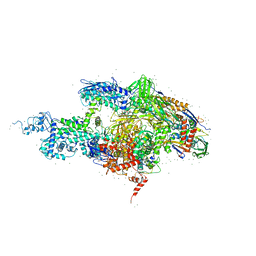 | | Crystal structure of the T. thermophilus RNA polymerase holoenzyme in complex with antibiotic rifabutin | | Descriptor: | DNA-directed RNA polymerase alpha chain, DNA-directed RNA polymerase beta chain, DNA-directed RNA polymerase beta' chain, ... | | Authors: | Artsimovitch, I, Vassylyeva, M.N, Svetlov, D, Svetlov, V, Perederina, A, Igarashi, N, Matsugaki, N, Wakatsuki, S, Tahirov, T.H, Vassylyev, D.G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-01 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Allosteric modulation of the RNA polymerase catalytic reaction is an essential component of transcription control by rifamycins.
Cell(Cambridge,Mass.), 122, 2005
|
|
2CMM
 
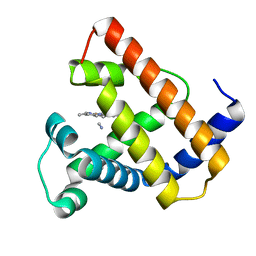 | | STRUCTURAL ANALYSIS OF THE MYOGLOBIN RECONSTITUTED WITH IRON PORPHINE | | Descriptor: | CYANIDE ION, MYOGLOBIN, PORPHYRIN FE(III) | | Authors: | Sato, T, Tanaka, N, Moriyama, H, Igarashi, N, Neya, S, Funasaki, N, Iizuka, T, Shiro, Y. | | Deposit date: | 1993-12-24 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis of the myoglobin reconstituted with iron porphine.
J.Biol.Chem., 268, 1993
|
|
2BE5
 
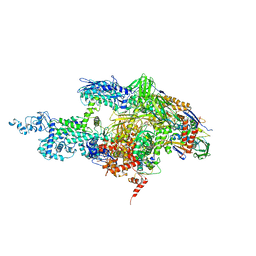 | | Crystal structure of the T. Thermophilus RNA polymerase holoenzyme in complex with inhibitor tagetitoxin | | Descriptor: | DNA-directed RNA polymerase alpha chain, DNA-directed RNA polymerase beta chain, DNA-directed RNA polymerase beta' chain, ... | | Authors: | Vassylyev, D.G, Svetlov, V, Vassylyeva, M.N, Perederina, A, Igarashi, N, Matsugaki, N, Wakatsuki, S, Artsimovitch, I, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-10-22 | | Release date: | 2005-11-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for transcription inhibition by tagetitoxin
Nat.Struct.Mol.Biol., 12, 2005
|
|
2Z5H
 
 | | Crystal structure of the head-to-tail junction of tropomyosin complexed with a fragment of TnT | | Descriptor: | General control protein GCN4 and Tropomyosin alpha-1 chain, Tropomyosin alpha-1 chain and General control protein GCN4, Troponin T, ... | | Authors: | Murakami, K, Nozawa, K, Tomii, K, Kudou, N, Igarashi, N, Shirakihara, Y, Wakatsuki, S, Stewart, M, Yasunaga, T, Wakabayashi, T. | | Deposit date: | 2007-07-12 | | Release date: | 2008-04-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Structural basis for tropomyosin overlap in thin (actin) filaments and the generation of a molecular swivel by troponin-T
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
2Z5I
 
 | | Crystal structure of the head-to-tail junction of tropomyosin | | Descriptor: | General control protein GCN4 and Tropomyosin alpha-1 chain, MAGNESIUM ION, Tropomyosin alpha-1 chain and General control protein GCN4 | | Authors: | Murakami, K, Nozawa, K, Tomii, K, Kudou, N, Igarashi, N, Shirakihara, Y, Wakatsuki, S, Stewart, M, Yasunaga, T, Wakabayashi, T. | | Deposit date: | 2007-07-12 | | Release date: | 2008-04-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for tropomyosin overlap in thin (actin) filaments and the generation of a molecular swivel by troponin-T
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
3W7Y
 
 | |
3W80
 
 | |
3AI4
 
 | | Crystal structure of yeast enhanced green fluorescent protein - mouse polymerase iota ubiquitin binding motif fusion protein | | Descriptor: | SULFATE ION, yeast enhanced green fluorescent protein,DNA polymerase iota | | Authors: | Suzuki, N, Wakatsuki, S, Kawasaki, M. | | Deposit date: | 2010-05-10 | | Release date: | 2010-09-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystallization of small proteins assisted by green fluorescent protein
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3AI5
 
 | | Crystal structure of yeast enhanced green fluorescent protein-ubiquitin fusion protein | | Descriptor: | 1,2-ETHANEDIOL, yeast enhanced green fluorescent protein,Ubiquitin | | Authors: | Suzuki, N, Wakatsuki, S, Kawasaki, M. | | Deposit date: | 2010-05-10 | | Release date: | 2010-09-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystallization of small proteins assisted by green fluorescent protein
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3W7Z
 
 | |
2A71
 
 | | Crystal structure of Emp47p carbohydrate recognition domain (CRD), orthorhombic crystal form | | Descriptor: | Emp47p | | Authors: | Satoh, T, Sato, K, Kanoh, A, Yamashita, K, Kato, R, Nakano, A, Wakatsuki, S. | | Deposit date: | 2005-07-04 | | Release date: | 2006-01-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structures of the carbohydrate recognition domain of Ca2+-independent cargo receptors Emp46p and Emp47p.
J.Biol.Chem., 281, 2006
|
|
2A6W
 
 | | Crystal structure of Emp46p carbohydrate recognition domain (CRD), metal-free form | | Descriptor: | Emp46p | | Authors: | Satoh, T, Sato, K, Kanoh, A, Yamashita, K, Kato, R, Nakano, A, Wakatsuki, S. | | Deposit date: | 2005-07-04 | | Release date: | 2006-01-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structures of the carbohydrate recognition domain of Ca2+-independent cargo receptors Emp46p and Emp47p.
J.Biol.Chem., 281, 2006
|
|
