2MS7
 
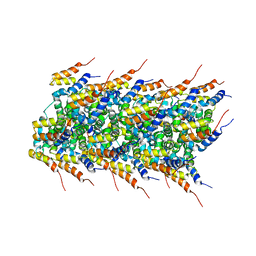 | | High-resolution solid-state NMR structure of the helical signal transduction filament MAVS CARD | | Descriptor: | Mitochondrial antiviral-signaling protein | | Authors: | He, L, Bardiaux, B, Spehr, J, Luehrs, T, Ritter, C. | | Deposit date: | 2014-07-25 | | Release date: | 2015-09-02 | | Last modified: | 2024-05-01 | | Method: | SOLID-STATE NMR | | Cite: | Structure determination of helical filaments by solid-state NMR spectroscopy.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
7DTO
 
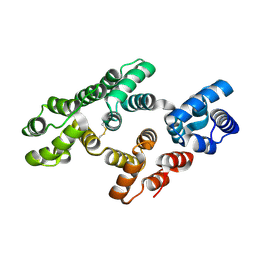 | |
7WLK
 
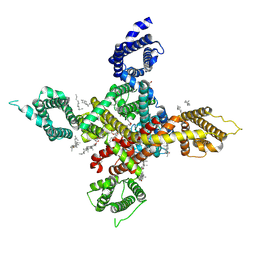 | | CryoEM structure of human low-voltage activated T-type calcium channel Cav3.3 in complex with Otilonium Bromide(OB) | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-[diethyl(methyl)-$l^{4}-azanyl]ethyl 4-[(2-octoxyphenyl)carbonylamino]benzoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | He, L, Yu, Z, Dong, Y, Chen, Q, Zhao, Y. | | Deposit date: | 2022-01-13 | | Release date: | 2022-05-04 | | Last modified: | 2022-05-25 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure, gating, and pharmacology of human Ca V 3.3 channel.
Nat Commun, 13, 2022
|
|
7WLI
 
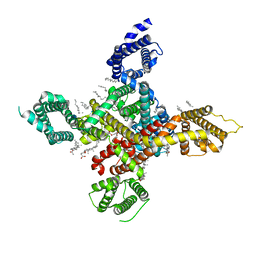 | | CryoEM structure of human low-voltage activated T-type calcium channel CaV3.3 (apo) | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | He, L, Yu, Z, Dong, Y, Chen, Q, Zhao, Y. | | Deposit date: | 2022-01-13 | | Release date: | 2022-05-04 | | Last modified: | 2022-05-25 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure, gating, and pharmacology of human Ca V 3.3 channel.
Nat Commun, 13, 2022
|
|
7WLJ
 
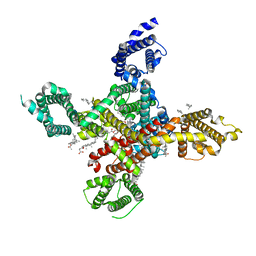 | | CryoEM structure of human low-voltage activated T-type calcium channel Cav3.3 in complex with mibefradil (MIB) | | Descriptor: | (1S,2S)-2-(2-{[3-(1H-benzimidazol-2-yl)propyl](methyl)amino}ethyl)-6-fluoro-1-(propan-2-yl)-1,2,3,4-tetrahydronaphthalen-2-yl methoxyacetate, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | He, L, Yu, Z, Dong, Y, Chen, Q, Zhao, Y. | | Deposit date: | 2022-01-13 | | Release date: | 2022-05-04 | | Last modified: | 2022-05-25 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure, gating, and pharmacology of human Ca V 3.3 channel.
Nat Commun, 13, 2022
|
|
7WLL
 
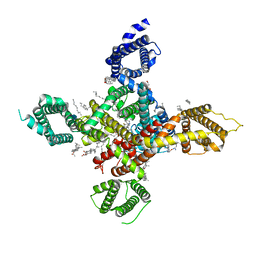 | | CryoEM structure of human low-voltage activated T-type calcium channel Cav3.3 in complex with pimozide(PMZ) | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-[1-[4,4-bis(4-fluorophenyl)butyl]piperidin-4-yl]-1~{H}-benzimidazol-2-one, ... | | Authors: | He, L, Yu, Z, Dong, Y, Chen, Q, Zhao, Y. | | Deposit date: | 2022-01-13 | | Release date: | 2022-05-04 | | Last modified: | 2022-05-25 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure, gating, and pharmacology of human Ca V 3.3 channel.
Nat Commun, 13, 2022
|
|
7XCE
 
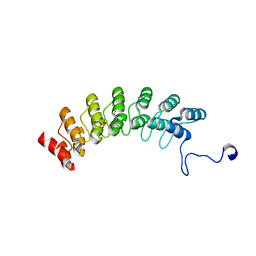 | | Crystal structure of Ankyrin G in complex with neurofascin | | Descriptor: | GLYCEROL, Neurofascin,Ankyrin-3, SULFATE ION | | Authors: | He, L, Li, J, Wang, C. | | Deposit date: | 2022-03-24 | | Release date: | 2022-09-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of Ankyrin-G in complex with a fragment of Neurofascin reveals binding mechanisms required for integrity of the axon initial segment.
J.Biol.Chem., 298, 2022
|
|
3DP1
 
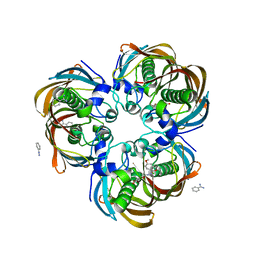 | | Crystal structure of (3R)-Hydroxyacyl-Acyl Carrier Protein Dehydratase (FabZ) from Helicobacter pylori in complex with compound 3n | | Descriptor: | (3R)-hydroxymyristoyl-acyl carrier protein dehydratase, BENZAMIDINE, CHLORIDE ION, ... | | Authors: | Zhang, L, He, L, Liu, X, Liu, H, Shen, X, Jiang, H. | | Deposit date: | 2008-07-07 | | Release date: | 2009-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovering potent inhibitors against the beta-hydroxyacyl-acyl carrier protein dehydratase (FabZ) of Helicobacter pylori: structure-based design, synthesis, bioassay, and crystal structure determination.
J.Med.Chem., 52, 2009
|
|
3DP0
 
 | | Crystal structure of (3R)-Hydroxyacyl-Acyl Carrier Protein Dehydratase (FabZ) from Helicobacter pylori in complex with compound 3m | | Descriptor: | (3R)-hydroxymyristoyl-acyl carrier protein dehydratase, BENZAMIDINE, CHLORIDE ION, ... | | Authors: | Zhang, L, He, L, Liu, X, Liu, H, Shen, X, Jiang, H. | | Deposit date: | 2008-07-07 | | Release date: | 2009-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovering potent inhibitors against the beta-hydroxyacyl-acyl carrier protein dehydratase (FabZ) of Helicobacter pylori: structure-based design, synthesis, bioassay, and crystal structure determination.
J.Med.Chem., 52, 2009
|
|
3DP2
 
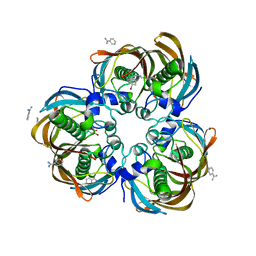 | | Crystal structure of (3R)-Hydroxyacyl-Acyl Carrier Protein Dehydratase (FabZ) from Helicobacter pylori in complex with compound 3j | | Descriptor: | (3R)-hydroxymyristoyl-acyl carrier protein dehydratase, 4-bromo-N'-[(1E)-(3,5-dibromo-2,4-dihydroxyphenyl)methylidene]benzohydrazide, BENZAMIDINE, ... | | Authors: | Zhang, L, He, L, Liu, X, Liu, H, Shen, X, Jiang, H. | | Deposit date: | 2008-07-07 | | Release date: | 2009-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovering potent inhibitors against the beta-hydroxyacyl-acyl carrier protein dehydratase (FabZ) of Helicobacter pylori: structure-based design, synthesis, bioassay, and crystal structure determination.
J.Med.Chem., 52, 2009
|
|
3DOY
 
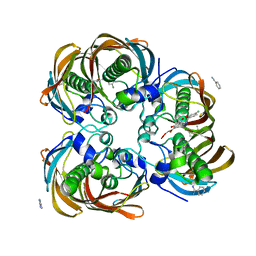 | | Crystal structure of (3R)-Hydroxyacyl-Acyl Carrier Protein Dehydratase (FabZ) from Helicobacter pylori in complex with compound 3i | | Descriptor: | (3R)-hydroxymyristoyl-acyl carrier protein dehydratase, 4-chloro-N'-[(1E)-(3,5-dibromo-2,4-dihydroxyphenyl)methylidene]benzohydrazide, BENZAMIDINE, ... | | Authors: | Zhang, L, He, L, Liu, X, Liu, H, Shen, X, Jiang, H. | | Deposit date: | 2008-07-07 | | Release date: | 2009-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovering potent inhibitors against the beta-hydroxyacyl-acyl carrier protein dehydratase (FabZ) of Helicobacter pylori: structure-based design, synthesis, bioassay, and crystal structure determination.
J.Med.Chem., 52, 2009
|
|
3DP3
 
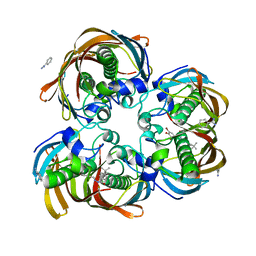 | | Crystal structure of (3R)-Hydroxyacyl-Acyl Carrier Protein Dehydratase (FabZ) from Helicobacter pylori in complex with compound 3q | | Descriptor: | (3R)-hydroxymyristoyl-acyl carrier protein dehydratase, 4-tert-butyl-N'-[(1E)-(3,5-dibromo-2,4-dihydroxyphenyl)methylidene]benzohydrazide, BENZAMIDINE, ... | | Authors: | Zhang, L, He, L, Liu, X, Liu, H, Shen, X, Jiang, H. | | Deposit date: | 2008-07-07 | | Release date: | 2009-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovering potent inhibitors against the beta-hydroxyacyl-acyl carrier protein dehydratase (FabZ) of Helicobacter pylori: structure-based design, synthesis, bioassay, and crystal structure determination.
J.Med.Chem., 52, 2009
|
|
3DOZ
 
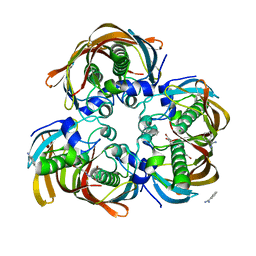 | | Crystal structure of (3R)-Hydroxyacyl-Acyl Carrier Protein Dehydratase (FabZ) from Helicobacter pylori in complex with compound 3k | | Descriptor: | (3R)-hydroxymyristoyl-acyl carrier protein dehydratase, 3-bromo-N'-[(1E)-(3,5-dibromo-2,4-dihydroxyphenyl)methylidene]benzohydrazide, BENZAMIDINE, ... | | Authors: | Zhang, L, He, L, Liu, X, Liu, H, Shen, X, Jiang, H. | | Deposit date: | 2008-07-07 | | Release date: | 2009-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovering potent inhibitors against the beta-hydroxyacyl-acyl carrier protein dehydratase (FabZ) of Helicobacter pylori: structure-based design, synthesis, bioassay, and crystal structure determination.
J.Med.Chem., 52, 2009
|
|
7YS6
 
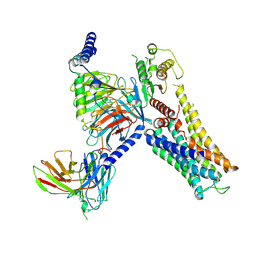 | | Cryo-EM structure of the Serotonin 6 (5-HT6) receptor-DNGs-scFv16 complex | | Descriptor: | 5-hydroxytryptamine receptor 6, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhao, Q.Y, Wang, Y.F, He, L, Wang, S, Cong, Y. | | Deposit date: | 2022-08-11 | | Release date: | 2023-03-29 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into constitutive activity of 5-HT 6 receptor.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
2MS8
 
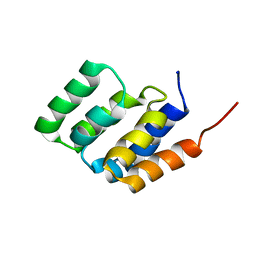 | | Solution NMR structure of MAVS CARD | | Descriptor: | Mitochondrial antiviral-signaling protein | | Authors: | Spehr, J, He, L, Luehrs, T, Ritter, C. | | Deposit date: | 2014-07-25 | | Release date: | 2015-09-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure determination of helical filaments by solid-state NMR spectroscopy.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
1QET
 
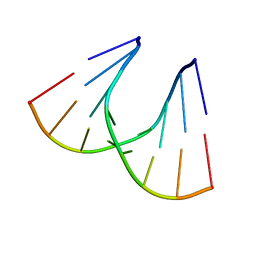 | | TANDEM GU MISMATCHES IN RNA, NMR, 30 STRUCTURES | | Descriptor: | RNA (5'-R(*GP*GP*AP*UP*GP*UP*CP*C)-3') | | Authors: | Mcdowell, J.A, He, L, Chen, X, Turner, D.H. | | Deposit date: | 1997-03-04 | | Release date: | 1997-06-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Investigation of the structural basis for thermodynamic stabilities of tandem GU wobble pairs: NMR structures of (rGGAGUUCC)2 and (rGGAUGUCC)2.
Biochemistry, 36, 1997
|
|
6GJQ
 
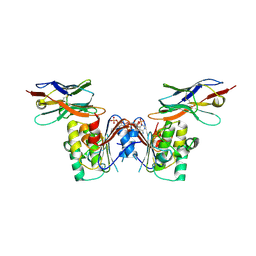 | | human NBD1 of CFTR in complex with nanobody T27 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cystic fibrosis transmembrane conductance regulator, Nanobody T27 | | Authors: | Sigoillot, M, Overtus, M, Grodecka, M, Scholl, D, Garcia-Pino, A, Laeremans, T, He, L, Pardon, E, Hildebrandt, E, Urbatsch, I, Steyaert, J, Riordan, J.R, Govaerts, C. | | Deposit date: | 2018-05-16 | | Release date: | 2019-06-19 | | Last modified: | 2019-08-21 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Domain-interface dynamics of CFTR revealed by stabilizing nanobodies.
Nat Commun, 10, 2019
|
|
6GKD
 
 | | human NBD1 of CFTR in complex with nanobodies D12 and G3a | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cystic fibrosis transmembrane conductance regulator, GLYCEROL, ... | | Authors: | Sigoillot, M, Overtus, M, Grodecka, M, Scholl, D, Garcia-Pino, A, Laeremans, T, He, L, Pardon, E, Hildebrandt, E, Urbatsch, I, Steyaert, J, Riordan, J.R, Govaerts, C. | | Deposit date: | 2018-05-18 | | Release date: | 2019-06-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Domain-interface dynamics of CFTR revealed by stabilizing nanobodies.
Nat Commun, 10, 2019
|
|
6GJS
 
 | | Human NBD1 of CFTR in complex with nanobodies D12 and T4 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cystic fibrosis transmembrane conductance regulator, MAGNESIUM ION, ... | | Authors: | Sigoillot, M, Overtus, M, Grodecka, M, Scholl, D, Garcia-Pino, A, Laeremans, T, He, L, Pardon, E, Hildebrandt, E, Urbatsch, I, Steyaert, J, Riordan, J.R, Govaerts, C. | | Deposit date: | 2018-05-16 | | Release date: | 2019-06-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Domain-interface dynamics of CFTR revealed by stabilizing nanobodies.
Nat Commun, 10, 2019
|
|
6GK4
 
 | | Human NBD1 of CFTR in complex with nanobodies D12 and T8 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cystic fibrosis transmembrane conductance regulator, GLYCEROL, ... | | Authors: | Sigoillot, M, Overtus, M, Grodecka, M, Scholl, D, Garcia-Pino, A, Laeremans, T, He, L, Pardon, E, Hildebrandt, E, Urbatsch, I, Steyaert, J, Riordan, J.R, Govaerts, C. | | Deposit date: | 2018-05-18 | | Release date: | 2019-06-19 | | Last modified: | 2019-08-21 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Domain-interface dynamics of CFTR revealed by stabilizing nanobodies.
Nat Commun, 10, 2019
|
|
6GJU
 
 | | human NBD1 of CFTR in complex with nanobodies T2a and T4 | | Descriptor: | Cystic fibrosis transmembrane conductance regulator, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Sigoillot, M, Overtus, M, Grodecka, M, Scholl, D, Garcia-Pino, A, Laeremans, T, He, L, Pardon, E, Hildebrandt, E, Urbatsch, I, Steyaert, J, Riordan, J.R, Govaerts, C. | | Deposit date: | 2018-05-17 | | Release date: | 2019-06-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Domain-interface dynamics of CFTR revealed by stabilizing nanobodies.
Nat Commun, 10, 2019
|
|
1QES
 
 | | TANDEM GU MISMATCHES IN RNA, NMR, 30 STRUCTURES | | Descriptor: | RNA (5'-R(*GP*GP*AP*GP*UP*UP*CP*C)-3') | | Authors: | Mcdowell, J.A, He, L, Chen, X, Turner, D.H. | | Deposit date: | 1997-03-04 | | Release date: | 1997-06-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Investigation of the structural basis for thermodynamic stabilities of tandem GU wobble pairs: NMR structures of (rGGAGUUCC)2 and (rGGAUGUCC)2.
Biochemistry, 36, 1997
|
|
5YEI
 
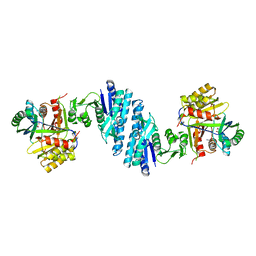 | | Mechanistic insight into the regulation of Pseudomonas aeruginosa aspartate kinase | | Descriptor: | Aspartokinase, GLYCEROL, LYSINE, ... | | Authors: | Li, C, Yang, M, Liu, L, Peng, C, Li, T, He, L, Song, Y, Zhu, Y, Bao, R. | | Deposit date: | 2017-09-17 | | Release date: | 2018-08-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Mechanistic insights into the allosteric regulation of Pseudomonas aeruginosa aspartate kinase.
Biochem.J., 475, 2018
|
|
5YQZ
 
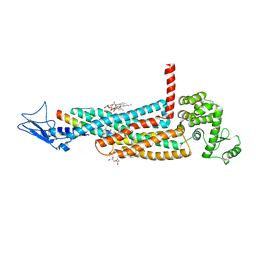 | | Structure of the glucagon receptor in complex with a glucagon analogue | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glucagon analogue, ... | | Authors: | Zhang, H, Qiao, A, Yang, L, VAN EPS, N, Frederiksen, K, Yang, D, Dai, A, Cai, X, Zhang, H, Yi, C, Can, C, He, L, Yang, H, Lau, J, Ernst, O, Hanson, M, Stevens, R, Wang, M, Seedtz-Runge, S, Jiang, H, Zhao, Q, Wu, B. | | Deposit date: | 2017-11-08 | | Release date: | 2018-01-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the glucagon receptor in complex with a glucagon analogue.
Nature, 553, 2018
|
|
6LKC
 
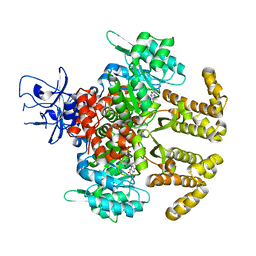 | | Crystal structure of PfaD from Shewanella piezotolerans in complex with FMN | | Descriptor: | CALCIUM ION, FLAVIN MONONUCLEOTIDE, GLYCEROL, ... | | Authors: | Zhang, M.L, Li, Q, Meng, S.S, Guo, L.J, He, L, Huang, J.Z, Li, L, Zhang, H.D. | | Deposit date: | 2019-12-19 | | Release date: | 2020-12-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | Structural Insights into the Trans -Acting Enoyl Reductase in the Biosynthesis of Long-Chain Polyunsaturated Fatty Acids in Shewanella piezotolerans .
J.Agric.Food Chem., 69, 2021
|
|
