2DGL
 
 | | Crystal structure of Escherichia coli GadB in complex with bromide | | Descriptor: | ACETIC ACID, BROMIDE ION, Glutamate decarboxylase beta, ... | | Authors: | Gruetter, M.G, Capitani, G, Gut, H. | | Deposit date: | 2006-03-14 | | Release date: | 2006-06-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Escherichia coli acid resistance: pH-sensing, activation by chloride and autoinhibition in GadB
Embo J., 25, 2006
|
|
2DGM
 
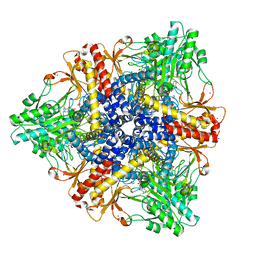 | | Crystal structure of Escherichia coli GadB in complex with iodide | | Descriptor: | ACETIC ACID, DI(HYDROXYETHYL)ETHER, FORMIC ACID, ... | | Authors: | Gruetter, M.G, Capitani, G, Gut, H. | | Deposit date: | 2006-03-14 | | Release date: | 2006-06-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Escherichia coli acid resistance: pH-sensing, activation by chloride and autoinhibition in GadB
Embo J., 25, 2006
|
|
2DGK
 
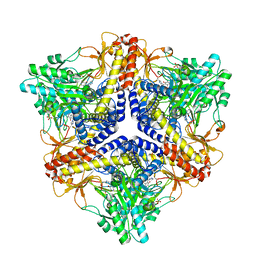 | | Crystal structure of an N-terminal deletion mutant of Escherichia coli GadB in an autoinhibited state (aldamine) | | Descriptor: | 1,2-ETHANEDIOL, Glutamate decarboxylase beta, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Gruetter, M.G, Capitani, G, Gut, H. | | Deposit date: | 2006-03-14 | | Release date: | 2006-06-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Escherichia coli acid resistance: pH-sensing, activation by chloride and autoinhibition in GadB
Embo J., 25, 2006
|
|
1AIE
 
 | |
4BS0
 
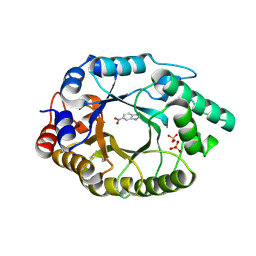 | | Crystal Structure of Kemp Eliminase HG3.17 E47N,N300D Complexed with Transition State Analog 6-Nitrobenzotriazole | | Descriptor: | 6-NITROBENZOTRIAZOLE, KEMP ELIMINASE HG3.17, SULFATE ION | | Authors: | Blomberg, R, Kries, H, Pinkas, D.M, Mittl, P.R.E, Gruetter, M.G, Privett, H.K, Mayo, S, Hilvert, D. | | Deposit date: | 2013-06-06 | | Release date: | 2013-10-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Precision is Essential for Efficient Catalysis in an Evolved Kemp Eliminase
Nature, 503, 2013
|
|
4CDL
 
 | | Crystal Structure of Retro-aldolase RA110.4-6 Complexed with Inhibitor 1-(6-methoxy-2-naphthalenyl)-1,3-butanedione | | Descriptor: | (2E)-1-(6-methoxynaphthalen-2-yl)but-2-en-1-one, STEROID DELTA-ISOMERASE | | Authors: | Pinkas, D.M, Studer, S, Obexer, R, Giger, L, Gruetter, M.G, Baker, D, Hilvert, D. | | Deposit date: | 2013-11-01 | | Release date: | 2014-11-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Active Site Plasticity of a Computationally Designed Retro-Aldolase Enzyme
To be Published
|
|
4CG4
 
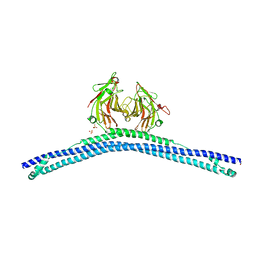 | | Crystal structure of the CHS-B30.2 domains of TRIM20 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-ETHANETHIOL, PYRIN, ... | | Authors: | Weinert, C, Morger, D, Djekic, A, Mittl, P.R.E, Gruetter, M.G. | | Deposit date: | 2013-11-20 | | Release date: | 2015-04-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Trim20 C-Terminal Coiled-Coil/B30.2 Fragment: Implications for the Recognition of Higher Order Oligomers
Sci.Rep., 5, 2015
|
|
2I1B
 
 | |
2FBE
 
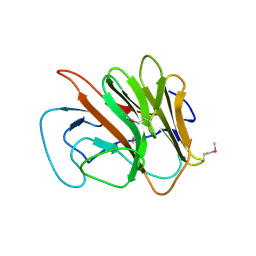 | | Crystal Structure of the PRYSPRY-domain | | Descriptor: | PREDICTED: similar to ret finger protein-like 1 | | Authors: | Gruetter, C, Briand, C, Capitani, G, Mittl, P.R, Gruetter, M.G. | | Deposit date: | 2005-12-09 | | Release date: | 2006-01-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Structure of the PRYSPRY-domain: Implications for autoinflammatory diseases
Febs Lett., 580, 2006
|
|
1VRS
 
 | | Crystal structure of the disulfide-linked complex between the N-terminal and C-terminal domain of the electron transfer catalyst DsbD | | Descriptor: | Thiol:disulfide interchange protein dsbD | | Authors: | Rozhkova, A, Stirnimann, C.U, Frei, P, Grauschopf, U, Brunisholz, R, Gruetter, M.G, Capitani, G, Glockshuber, R. | | Deposit date: | 2005-06-17 | | Release date: | 2005-07-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural basis and kinetics of inter- and intramolecular disulfide exchange in the redox catalyst DsbD
Embo J., 23, 2004
|
|
1Z5Y
 
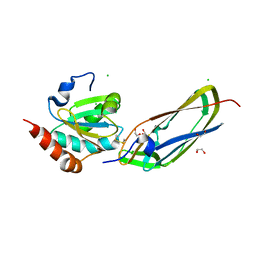 | | Crystal Structure Of The Disulfide-Linked Complex Between The N-Terminal Domain Of The Electron Transfer Catalyst DsbD and The Cytochrome c Biogenesis Protein CcmG | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Thiol:disulfide interchange protein dsbD, ... | | Authors: | Stirnimann, C.U, Rozhkova, A, Grauschopf, U, Gruetter, M.G, Glockshuber, R, Capitani, G. | | Deposit date: | 2005-03-21 | | Release date: | 2005-07-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural Basis and Kinetics of DsbD-Dependent Cytochrome c Maturation
STRUCTURE, 13, 2005
|
|
2WL1
 
 | | Pyrin PrySpry domain | | Descriptor: | 1,2-ETHANEDIOL, PYRIN, THIOCYANATE ION | | Authors: | Weinert, C, Mittl, P.R, Gruetter, M.G. | | Deposit date: | 2009-06-19 | | Release date: | 2009-10-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | The Crystal Structure of Human Pyrin B30.2 Domain: Implications for Mutations Associated with Familial Mediterranean Fever.
J.Mol.Biol., 394, 2009
|
|
2FWG
 
 | | high resolution crystal structure of the C-terminal domain of the electron transfer catalyst DsbD (photoreduced form) | | Descriptor: | Thiol:disulfide interchange protein dsbD | | Authors: | Stirnimann, C.U, Rozhkova, A, Grauschopf, U, Boeckmann, R.A, Glockshuber, R, Capitani, G, Gruetter, M.G. | | Deposit date: | 2006-02-02 | | Release date: | 2006-06-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | High-resolution structures of Escherichia coli cDsbD in different redox states: A combined crystallographic, biochemical and computational study
J.Mol.Biol., 358, 2006
|
|
2FWE
 
 | | crystal structure of the C-terminal domain of the electron transfer catalyst DsbD (oxidized form) | | Descriptor: | IODIDE ION, NICKEL (II) ION, SODIUM ION, ... | | Authors: | Stirnimann, C.U, Rozhkova, A, Grauschopf, U, Boeckmann, R.A, Glockshuber, R, Capitani, G, Gruetter, M.G. | | Deposit date: | 2006-02-02 | | Release date: | 2006-06-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | High-resolution structures of Escherichia coli cDsbD in different redox states: A combined crystallographic, biochemical and computational study
J.Mol.Biol., 358, 2006
|
|
2FWF
 
 | | high resolution crystal structure of the C-terminal domain of the electron transfer catalyst DsbD (reduced form) | | Descriptor: | IODIDE ION, SODIUM ION, Thiol:disulfide interchange protein dsbD | | Authors: | Stirnimann, C.U, Rozhkova, A, Grauschopf, U, Boeckmann, R.A, Glockshuber, R, Capitani, G, Gruetter, M.G. | | Deposit date: | 2006-02-02 | | Release date: | 2006-06-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | High-resolution structures of Escherichia coli cDsbD in different redox states: A combined crystallographic, biochemical and computational study
J.Mol.Biol., 358, 2006
|
|
2FWH
 
 | | atomic resolution crystal structure of the C-terminal domain of the electron transfer catalyst DsbD (reduced form at pH7) | | Descriptor: | DI(HYDROXYETHYL)ETHER, IODIDE ION, Thiol:disulfide interchange protein dsbD | | Authors: | Stirnimann, C.U, Rozhkova, A, Grauschopf, U, Boeckmann, R.A, Glockshuber, R, Capitani, G, Gruetter, M.G. | | Deposit date: | 2006-02-02 | | Release date: | 2006-06-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (0.99 Å) | | Cite: | High-resolution structures of Escherichia coli cDsbD in different redox states: A combined crystallographic, biochemical and computational study
J.Mol.Biol., 358, 2006
|
|
1M7Y
 
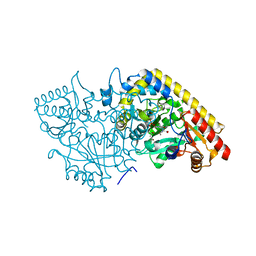 | | Crystal structure of apple ACC synthase in complex with L-aminoethoxyvinylglycine | | Descriptor: | (2E,3E)-4-(2-aminoethoxy)-2-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)imino]but-3-enoic acid, (4R)-2-METHYLPENTANE-2,4-DIOL, 1-aminocyclopropane-1-carboxylate synthase | | Authors: | Capitani, G, McCarthy, D, Gut, H, Gruetter, M.G, Kirsch, J.F. | | Deposit date: | 2002-07-23 | | Release date: | 2002-12-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Apple 1-Aminocyclopropane-1-carboxylate Synthase in Complex with the Inhibitor
L-Aminoethoxyvinylglycine
J.Biol.Chem., 277, 2002
|
|
1MJ0
 
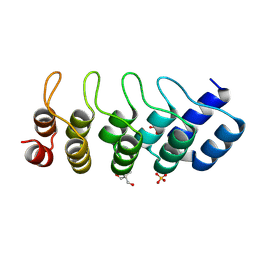 | | SANK E3_5: an artificial Ankyrin repeat protein | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, SANK E3_5 Protein, SULFATE ION | | Authors: | Kohl, A, Binz, H.K, Forrer, P, Stumpp, M.T, Plueckthun, A, Gruetter, M.G. | | Deposit date: | 2002-08-26 | | Release date: | 2003-01-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.031 Å) | | Cite: | Designed to be stable: Crystal structure of a consensus ankyrin repeat protein
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1TMT
 
 | |
1TMU
 
 | |
2RKO
 
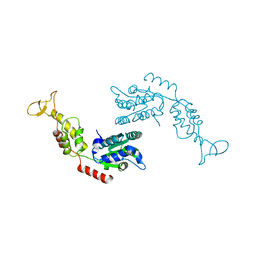 | | Crystal Structure of the Vps4p-dimer | | Descriptor: | Vacuolar protein sorting-associated protein 4 | | Authors: | Hartmann, C, Gruetter, M.G. | | Deposit date: | 2007-10-17 | | Release date: | 2008-02-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Vacuolar protein sorting: two different functional states of the AAA-ATPase Vps4p
J.Mol.Biol., 377, 2008
|
|
2V7N
 
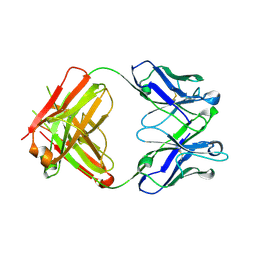 | | Unusual twinning in crystals of the CitS binding antibody Fab fragment f3p4 | | Descriptor: | IMMUNOGLOBULIN HEAVY CHAIN, IMMUNOGLOBULIN LIGHT CHAIN | | Authors: | Frey, D, Huber, T, Plueckthun, A, Gruetter, M.G. | | Deposit date: | 2007-07-31 | | Release date: | 2008-06-17 | | Last modified: | 2019-05-22 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structure of the Recombinant Antibody Fab Fragment F3P4.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
2V50
 
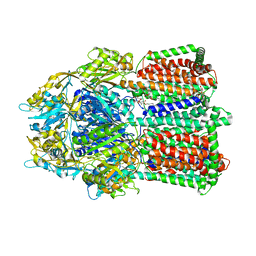 | |
3NFB
 
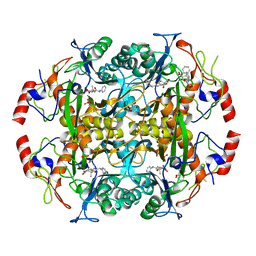 | | Crystal structure of the N-terminal beta-aminopeptidase BapA in complex with hydrolyzed ampicillin | | Descriptor: | (2S,4S)-2-[(R)-{[(2R)-2-amino-2-phenylacetyl]amino}(carboxy)methyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, Beta-peptidyl aminopeptidase, GLYCEROL, ... | | Authors: | Merz, T, Heck, T, Geueke, B, Kohler, H.-P.E, Gruetter, M.G. | | Deposit date: | 2010-06-10 | | Release date: | 2011-09-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure and inhibition of the beta-aminopeptidase BapA, a new ampicillin-recognizing member of the N-terminal nucleophile hydrolase family
To be Published
|
|
3NDV
 
 | | Crystal structure of the N-terminal beta-aminopeptidase BapA in complex with ampicillin | | Descriptor: | (2S,4S)-2-[(1R)-1-{[(2R)-2-amino-2-phenylacetyl]amino}-2-oxoethyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, (2S,5R,6R)-6-{[(2R)-2-AMINO-2-PHENYLETHANOYL]AMINO}-3,3-DIMETHYL-7-OXO-4-THIA-1-AZABICYCLO[3.2.0]HEPTANE-2-CARBOXYLIC ACID, Beta-peptidyl aminopeptidase, ... | | Authors: | Merz, T, Heck, T, Geueke, B, Kohler, H.-P.E, Gruetter, M.G. | | Deposit date: | 2010-06-08 | | Release date: | 2011-09-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures and inhibition of the beta-aminopeptidase BapA, a new ampicillin-recognizing member of the N-terminal nucleophile hydrolase family
To be Published
|
|
