3S0R
 
 | |
1XOF
 
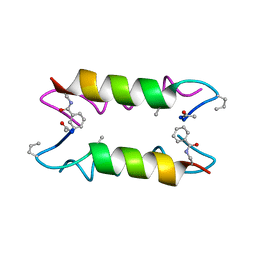 | | Heterooligomeric Beta Beta Alpha Miniprotein | | Descriptor: | BBAhetT1 | | Authors: | Ali, M.H, Taylor, C.M, Grigoryan, G, Allen, K.N, Imperiali, B, Keating, A.E. | | Deposit date: | 2004-10-06 | | Release date: | 2005-02-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Design of a Heterospecific, Tetrameric, 21-Residue Miniprotein with Mixed alpha/beta Structure.
Structure, 13, 2005
|
|
5ET3
 
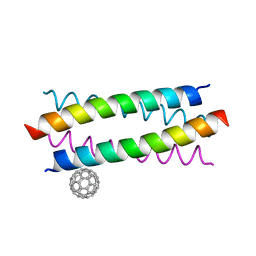 | | Crystal Structure of De novo Designed Fullerene organizing peptide | | Descriptor: | (C_{60}-I_{h})[5,6]fullerene, Fullerene Organizing Protein (C60Sol-COP-3) | | Authors: | Kim, K.-H, Kim, Y.H, Acharya, R, Kim, N.H, Paul, J, Grigoryan, G, DeGrado, W.F. | | Deposit date: | 2015-11-17 | | Release date: | 2016-05-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.671 Å) | | Cite: | Protein-directed self-assembly of a fullerene crystal.
Nat Commun, 7, 2016
|
|
5HKN
 
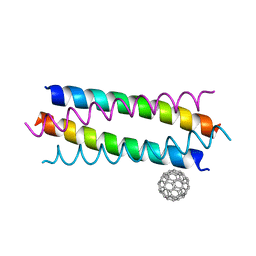 | | Crystal structure de novo designed fullerene organizing protein complex with fullerene | | Descriptor: | (C_{60}-I_{h})[5,6]fullerene, fullerene organizing protein | | Authors: | Paul, J, Acharya, R, Kim, K.-H, Kim, Y.H, Kim, N.H, Grigoryan, G, DeGardo, W.F. | | Deposit date: | 2016-01-14 | | Release date: | 2016-05-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.761 Å) | | Cite: | Protein-directed self-assembly of a fullerene crystal.
Nat Commun, 7, 2016
|
|
5HKR
 
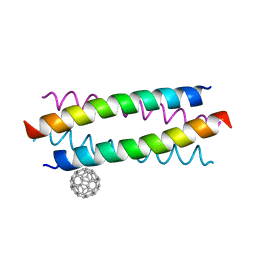 | | Crystal structure of de novo designed fullerene organising protein complex with fullerene | | Descriptor: | (C_{60}-I_{h})[5,6]fullerene, fullerene organizing protein | | Authors: | Acharya, R, Kim, Y.H, Grigoryan, G, DeGardo, W.F. | | Deposit date: | 2016-01-14 | | Release date: | 2016-05-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Protein-directed self-assembly of a fullerene crystal.
Nat Commun, 7, 2016
|
|
8TNO
 
 | |
8TNM
 
 | |
7KUW
 
 | |
2MUZ
 
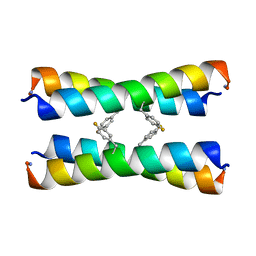 | | ssNMR structure of a designed rocker protein | | Descriptor: | designed rocker protein | | Authors: | Wang, T, Joh, N, Wu, Y, DeGrado, W.F, Hong, M. | | Deposit date: | 2014-09-18 | | Release date: | 2014-12-24 | | Last modified: | 2015-01-14 | | Method: | SOLUTION NMR | | Cite: | De novo design of a transmembrane Zn2+-transporting four-helix bundle.
Science, 346, 2014
|
|
4P6J
 
 | |
4P6K
 
 | |
4P6L
 
 | |
6MBB
 
 | | Human Bfl-1 in complex with the designed peptide dF1 | | Descriptor: | Bcl-2-related protein A1, dF1 | | Authors: | Jenson, J.M, Keating, A.E. | | Deposit date: | 2018-08-29 | | Release date: | 2019-03-06 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Tertiary Structural Motif Sequence Statistics Enable Facile Prediction and Design of Peptides that Bind Anti-apoptotic Bfl-1 and Mcl-1.
Structure, 27, 2019
|
|
6MBE
 
 | | Human Mcl-1 in complex with the designed peptide dM7 | | Descriptor: | CHLORIDE ION, Induced myeloid leukemia cell differentiation protein Mcl-1, dM7 | | Authors: | Jenson, J.M, Keating, A.E. | | Deposit date: | 2018-08-29 | | Release date: | 2019-03-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Tertiary Structural Motif Sequence Statistics Enable Facile Prediction and Design of Peptides that Bind Anti-apoptotic Bfl-1 and Mcl-1.
Structure, 27, 2019
|
|
6MBD
 
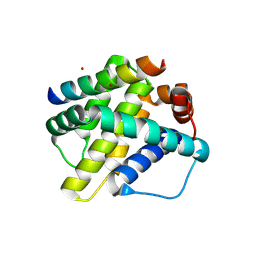 | | Human Mcl-1 in complex with the designed peptide dM1 | | Descriptor: | Induced myeloid leukemia cell differentiation protein Mcl-1, ZINC ION, dM1 | | Authors: | Jenson, J.M, Keating, A.E. | | Deposit date: | 2018-08-29 | | Release date: | 2019-03-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Tertiary Structural Motif Sequence Statistics Enable Facile Prediction and Design of Peptides that Bind Anti-apoptotic Bfl-1 and Mcl-1.
Structure, 27, 2019
|
|
6MBC
 
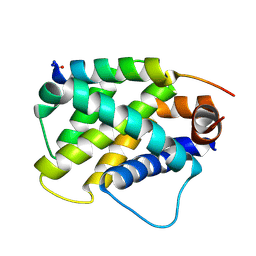 | | Human Bfl-1 in complex with the designed peptide dF4 | | Descriptor: | Bcl-2-related protein A1, dF4 | | Authors: | Jenson, J.M, Keating, A.E. | | Deposit date: | 2018-08-29 | | Release date: | 2019-03-06 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (1.752 Å) | | Cite: | Tertiary Structural Motif Sequence Statistics Enable Facile Prediction and Design of Peptides that Bind Anti-apoptotic Bfl-1 and Mcl-1.
Structure, 27, 2019
|
|
6WL7
 
 | | Cryo-EM of Form 2 like peptide filament, 29-20-2 | | Descriptor: | peptide 29-20-2 | | Authors: | Wang, F, Gnewou, O.M, Modlin, C, Egelman, E.H, Conticello, V.P. | | Deposit date: | 2020-04-18 | | Release date: | 2020-12-02 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural analysis of cross alpha-helical nanotubes provides insight into the designability of filamentous peptide nanomaterials.
Nat Commun, 12, 2021
|
|
6WKX
 
 | | Cryo-EM of Form 1 related peptide filament, 15-10-3 | | Descriptor: | peptide 15-10-3 | | Authors: | Wang, F, Gnewou, O.M, Egelman, E.H, Conticello, V.P. | | Deposit date: | 2020-04-17 | | Release date: | 2020-12-02 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural analysis of cross alpha-helical nanotubes provides insight into the designability of filamentous peptide nanomaterials.
Nat Commun, 12, 2021
|
|
6WL1
 
 | | Cryo-EM of Form 1 related peptide filament, 36-31-3 | | Descriptor: | peptide 36-31-3 | | Authors: | Wang, F, Gnewou, O.M, Modlin, C, Egelman, E.H, Conticello, V.P. | | Deposit date: | 2020-04-17 | | Release date: | 2020-12-02 | | Last modified: | 2021-12-15 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural analysis of cross alpha-helical nanotubes provides insight into the designability of filamentous peptide nanomaterials.
Nat Commun, 12, 2021
|
|
6WL0
 
 | | Cryo-EM of Form 1 related peptide filament, 36-31-3-RD | | Descriptor: | peptide 36-31-3-RD | | Authors: | Wang, F, Gnewou, O.M, Su, Z, Egelman, E.H, Conticello, V.P. | | Deposit date: | 2020-04-17 | | Release date: | 2020-12-02 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structural analysis of cross alpha-helical nanotubes provides insight into the designability of filamentous peptide nanomaterials.
Nat Commun, 12, 2021
|
|
6WL8
 
 | | Cryo-EM of Form 2 peptide filament | | Descriptor: | Form 2 peptide | | Authors: | Wang, F, Gnewou, O.M, Xu, C, Su, Z, Egelman, E.H, Conticello, V.P. | | Deposit date: | 2020-04-18 | | Release date: | 2020-12-02 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural analysis of cross alpha-helical nanotubes provides insight into the designability of filamentous peptide nanomaterials.
Nat Commun, 12, 2021
|
|
6WL9
 
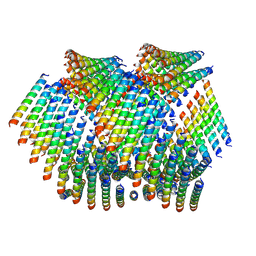 | | Cryo-EM of Form 2 like peptide filament, Form2a | | Descriptor: | peptide Form2a | | Authors: | Wang, F, Beltran, L.C, Gnewou, O.M, Egelman, E.H, Conticello, V.P. | | Deposit date: | 2020-04-18 | | Release date: | 2020-12-02 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural analysis of cross alpha-helical nanotubes provides insight into the designability of filamentous peptide nanomaterials.
Nat Commun, 12, 2021
|
|
6WKY
 
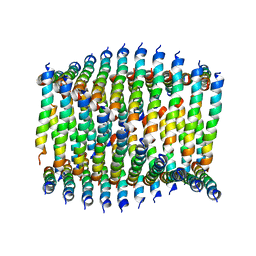 | | Cryo-EM of Form 1 related peptide filament, 29-24-3 | | Descriptor: | peptide 29-24-3 | | Authors: | Wang, F, Gnewou, O.M, Egelman, E.H, Conticello, V.P. | | Deposit date: | 2020-04-17 | | Release date: | 2020-12-02 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural analysis of cross alpha-helical nanotubes provides insight into the designability of filamentous peptide nanomaterials.
Nat Commun, 12, 2021
|
|
