4AMH
 
 | | Influence of circular permutation on the folding pathway of a PDZ domain | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DISKS LARGE HOMOLOG 1, GLYCEROL | | Authors: | Hultqvist, G, Punekar, A.S, Chi, C.N, Selmer, M, Gianni, S, Jemth, P. | | Deposit date: | 2012-03-10 | | Release date: | 2012-12-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Tolerance of Protein Folding to a Circular Permutation in a Pdz Domain
Plos One, 7, 2012
|
|
1YNR
 
 | | Crystal structure of the cytochrome c-552 from Hydrogenobacter thermophilus at 2.0 resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Cytochrome c-552, HEME C, ... | | Authors: | Travaglini-Allocatelli, C, Gianni, S, Dubey, V.K, Borgia, A, Di Matteo, A, Bonivento, D, Cutruzzola, F, Bren, K.L, Brunori, M. | | Deposit date: | 2005-01-25 | | Release date: | 2005-05-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | An Obligatory Intermediate in the Folding Pathway of Cytochrome c552 from Hydrogenobacter thermophilus
J.Biol.Chem., 280, 2005
|
|
3RBL
 
 | | Crystal structure of Human aromatic L-amino acid decarboxylase (AADC) in the apo form | | Descriptor: | CHLORIDE ION, aromatic L-amino acid decarboxylase | | Authors: | Giardina, G, Montioli, R, Gianni, S, Cellini, B, Paiardini, A, Borri Voltattorni, C, Cutruzzola, F. | | Deposit date: | 2011-03-29 | | Release date: | 2011-10-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | Open conformation of human DOPA decarboxylase reveals the mechanism of PLP addition to Group II decarboxylases.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3RCH
 
 | | Crystal structure of Human aromatic L-amino acid decarboxylase (AADC) in the open conformation with LLP and PLP bound to Chain-A and Chain-B respectively | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, aromatic L-amino acid decarboxylase | | Authors: | Giardina, G, Montioli, R, Gianni, S, Cellini, B, Paiardini, A, Borri Voltattorni, C, Cutruzzola, F. | | Deposit date: | 2011-03-31 | | Release date: | 2011-10-19 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Open conformation of human DOPA decarboxylase reveals the mechanism of PLP addition to Group II decarboxylases.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2WIO
 
 | | Structure of the histidine tagged, open cytochrome P450 Eryk from S. erythraea | | Descriptor: | ERYTHROMYCIN B/D C-12 HYDROXYLASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Savino, C, Montemiglio, L.C, Sciara, G, Miele, A.E, Kedrew, S.G, Gianni, S, Vallone, B. | | Deposit date: | 2009-05-14 | | Release date: | 2009-07-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Investigating the Structural Plasticity of a Cytochrome P450: Three-Dimensional Structures of P450 Eryk and Binding to its Physiological Substrate.
J.Biol.Chem., 284, 2009
|
|
2XFH
 
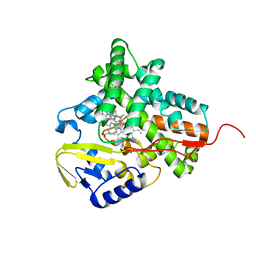 | | Structure of cytochrome P450 EryK cocrystallized with inhibitor clotrimazole. | | Descriptor: | 1-[(2-CHLOROPHENYL)(DIPHENYL)METHYL]-1H-IMIDAZOLE, DIMETHYL SULFOXIDE, ERYTHROMYCIN B/D C-12 HYDROXYLASE, ... | | Authors: | Savino, C, Montemiglio, L.C, Gianni, S, Vallone, B. | | Deposit date: | 2010-05-24 | | Release date: | 2010-09-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Azole Drugs Trap Cytochrome P450 Eryk in Alternative Conformational States.
Biochemistry, 49, 2010
|
|
3KH7
 
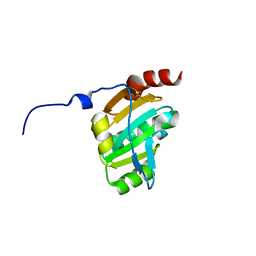 | | Crystal structure of the periplasmic soluble domain of reduced CcmG from Pseudomonas aeruginosa | | Descriptor: | Thiol:disulfide interchange protein dsbE | | Authors: | Di Matteo, A, Calosci, N, Gianni, S, Jemth, P, Brunori, M, Travaglini Allocatelli, C. | | Deposit date: | 2009-10-30 | | Release date: | 2010-04-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural and functional characterization of CcmG from Pseudomonas aeruginosa, a key component of the bacterial cytochrome c maturation apparatus.
Proteins, 78, 2010
|
|
3KH9
 
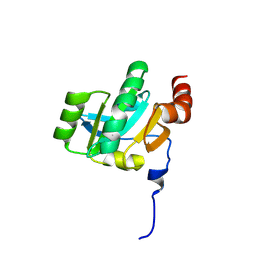 | | Crystal structure of the periplasmic soluble domain of oxidized CcmG from Pseudomonas aeruginosa | | Descriptor: | Thiol:disulfide interchange protein dsbE | | Authors: | Di Matteo, A, Calosci, N, Gianni, S, Jemth, P, Brunori, M, Travaglini Allocatelli, C. | | Deposit date: | 2009-10-30 | | Release date: | 2010-04-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and functional characterization of CcmG from Pseudomonas aeruginosa, a key component of the bacterial cytochrome c maturation apparatus.
Proteins, 78, 2010
|
|
3RBF
 
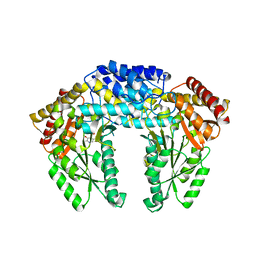 | | Crystal structure of Human aromatic L-amino acid decarboxylase (AADC) in the apo form | | Descriptor: | Aromatic-L-amino-acid decarboxylase, CHLORIDE ION, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Giardina, G, Montioli, R, Gianni, S, Cellini, B, Paiardini, A, Borri Voltattorni, C, Cutruzzola, F. | | Deposit date: | 2011-03-29 | | Release date: | 2011-10-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Open conformation of human DOPA decarboxylase reveals the mechanism of PLP addition to Group II decarboxylases.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2X7Z
 
 | | Crystal Structure of the SAP97 PDZ2 I342W C378A mutant protein domain | | Descriptor: | AMMONIUM ION, DISKS LARGE HOMOLOG 1, IMIDAZOLE | | Authors: | Haq, S.R, Jurgens, M.C, Chi, C.N, Elfstrom, L, Koh, C.S, Selmer, M, Gianni, S, Jemth, P. | | Deposit date: | 2010-03-04 | | Release date: | 2010-03-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Plastic Energy Landscape of Protein Folding: A Triangular Folding Mechanism with an Equilibrium Intermediate for a Small Protein Domain.
J.Biol.Chem., 285, 2010
|
|
2HL7
 
 | | Crystal structure of the periplasmic domain of CcmH from Pseudomonas aeruginosa | | Descriptor: | Cytochrome C-type biogenesis protein CcmH, TETRAETHYLENE GLYCOL | | Authors: | Di Matteo, A, Travaglini-Allocatelli, C, Gianni, S, Brunori, M. | | Deposit date: | 2006-07-06 | | Release date: | 2007-07-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A strategic protein in cytochrome c maturation: three-dimensional structure of CcmH and binding to apocytochrome c
J.Biol.Chem., 282, 2007
|
|
1VJ6
 
 | |
2NTO
 
 | | Structure of the Glutathione Transferase from Ochrobactrum anthropi in complex with glutathione | | Descriptor: | GLUTATHIONE, SULFATE ION, glutathione S-transferase | | Authors: | Federici, L, Bonivento, D, Di Matteo, A, Allocati, N. | | Deposit date: | 2006-11-08 | | Release date: | 2007-09-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.095 Å) | | Cite: | Role of Ser11 in the stabilization of the structure of Ochrobactrum anthropi glutathione transferase
Biochem.J., 403, 2007
|
|
2JJP
 
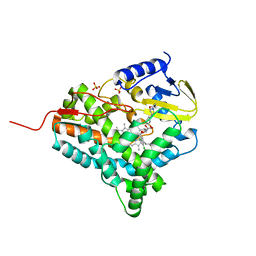 | | Structure of cytochrome P450 EryK in complex with inhibitor ketoconazole (KC) | | Descriptor: | 1-ACETYL-4-(4-{[(2S,4R)-2-(2,4-DICHLOROPHENYL)-2-(1H-IMIDAZOL-1-YLMETHYL)-1,3-DIOXOLAN-4-YL]METHOXY}PHENYL)PIPERAZINE, CYTOCHROME P450 113A1, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Savino, C, Sciara, G, Miele, A.E, Kendrew, S.G, Vallone, B. | | Deposit date: | 2008-04-15 | | Release date: | 2009-07-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Azole Drugs Trap Cytochrome P450 Eryk in Alternative Conformational States.
Biochemistry, 49, 2010
|
|
2JJO
 
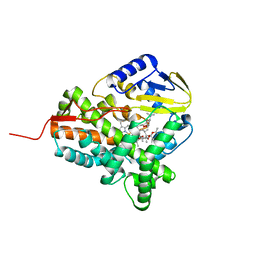 | | Structure of cytochrome P450 EryK in complex with its natural substrate erD | | Descriptor: | CYTOCHROME P450 113A1, Erythromycin D, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Savino, C, Sciara, G, Miele, A.E, Kendrew, S.G, Vallone, B. | | Deposit date: | 2008-04-15 | | Release date: | 2009-07-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Investigating the Structural Plasticity of a Cytochrome P450: Three-Dimensional Structures of P450 Eryk and Binding to its Physiological Substrate.
J.Biol.Chem., 284, 2009
|
|
2JJN
 
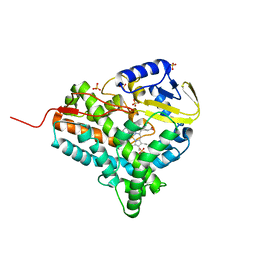 | | Structure of closed cytochrome P450 EryK | | Descriptor: | CYTOCHROME P450 113A1, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Savino, C, Sciara, G, Miele, A.E, Kendrew, S.G, Vallone, B. | | Deposit date: | 2008-04-15 | | Release date: | 2009-07-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Investigating the Structural Plasticity of a Cytochrome P450: Three-Dimensional Structures of P450 Eryk and Binding to its Physiological Substrate.
J.Biol.Chem., 284, 2009
|
|
2EXV
 
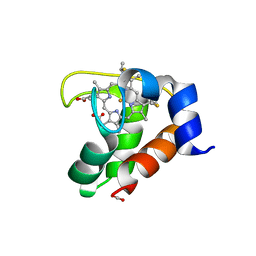 | | Crystal structure of the F7A mutant of the cytochrome c551 from Pseudomonas aeruginosa | | Descriptor: | ACETIC ACID, Cytochrome c-551, HEME C | | Authors: | Bonivento, D, Di Matteo, A, Borgia, A, Travaglini-Allocatelli, C, Brunori, M. | | Deposit date: | 2005-11-09 | | Release date: | 2006-02-07 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Unveiling a Hidden Folding Intermediate in c-Type Cytochromes by Protein Engineering
J.Biol.Chem., 281, 2006
|
|
