7P2O
 
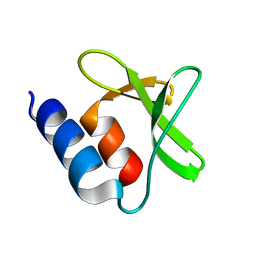 | |
5MTI
 
 | | Bamb_5917 Acyl-Carrier Protein | | Descriptor: | Phosphopantetheine-binding protein | | Authors: | Gallo, A, Kosol, S, Griffiths, D, Masschelein, J, Alkhalaf, L, Smith, H, Valentic, T, Tsai, S, Challis, G, Lewandowski, J.R. | | Deposit date: | 2017-01-09 | | Release date: | 2018-08-01 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural basis for chain release from the enacyloxin polyketide synthase
Nat.Chem., 2019
|
|
2LLH
 
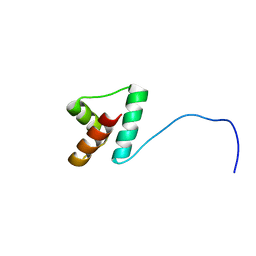 | | NMR structure of Npm1_c70 | | Descriptor: | Nucleophosmin | | Authors: | Banci, L, Bertini, I, Brunori, M, Di Matteo, A, Federici, L, Gallo, A, Lo Sterzo, C, Mori, M. | | Deposit date: | 2011-11-09 | | Release date: | 2012-06-27 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure of Nucleophosmin DNA-binding Domain and Analysis of Its Complex with a G-quadruplex Sequence from the c-MYC Promoter.
J.Biol.Chem., 287, 2012
|
|
2MJD
 
 | | Oxidized Yeast Adrenodoxin Homolog 1 | | Descriptor: | Adrenodoxin homolog, mitochondrial, FE2/S2 (INORGANIC) CLUSTER | | Authors: | Gallo, A, Banci, L. | | Deposit date: | 2014-01-07 | | Release date: | 2014-11-19 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Functional reconstitution of mitochondrial Fe/S cluster synthesis on Isu1 reveals the involvement of ferredoxin.
Nat Commun, 5, 2014
|
|
2MJE
 
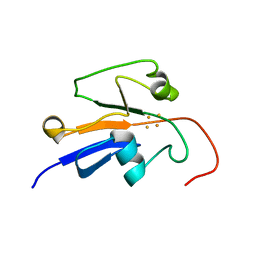 | | Reduced Yeast Adrenodoxin Homolog 1 | | Descriptor: | Adrenodoxin homolog, mitochondrial, FE2/S2 (INORGANIC) CLUSTER | | Authors: | Gallo, A, Banci, L. | | Deposit date: | 2014-01-07 | | Release date: | 2014-11-19 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Functional reconstitution of mitochondrial Fe/S cluster synthesis on Isu1 reveals the involvement of ferredoxin.
Nat Commun, 5, 2014
|
|
3O55
 
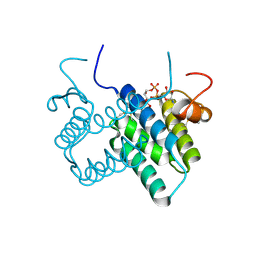 | | Crystal structure of human FAD-linked augmenter of liver regeneration (ALR) | | Descriptor: | Augmenter of liver regeneration, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Banci, L, Bertini, I, Calderone, V, Cefaro, C, Ciofi-Baffoni, S, Gallo, A, Kallergi, E, Lionaki, E, Pozidis, C, Tokatlidis, K. | | Deposit date: | 2010-07-28 | | Release date: | 2011-04-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular recognition and substrate mimicry drive the electron-transfer process between MIA40 and ALR.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3U2L
 
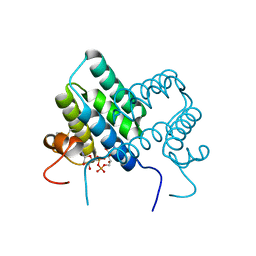 | | Crystal structure of human ALR mutant C142S. | | Descriptor: | FAD-linked sulfhydryl oxidase ALR, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Banci, L, Bertini, I, Calderone, V, Cefaro, C, Ciofi-Baffoni, S, Gallo, A. | | Deposit date: | 2011-10-04 | | Release date: | 2012-06-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | An electron-transfer path through an extended disulfide relay system: the case of the redox protein ALR.
J.Am.Chem.Soc., 134, 2012
|
|
3U2M
 
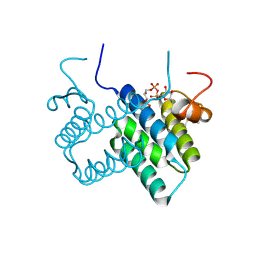 | | Crystal structure of human ALR mutant C142/145S | | Descriptor: | FAD-linked sulfhydryl oxidase ALR, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Banci, L, Bertini, I, Calderone, V, Cefaro, C, Ciofi-Baffoni, S, Gallo, A. | | Deposit date: | 2011-10-04 | | Release date: | 2012-06-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | An electron-transfer path through an extended disulfide relay system: the case of the redox protein ALR.
J.Am.Chem.Soc., 134, 2012
|
|
6CGO
 
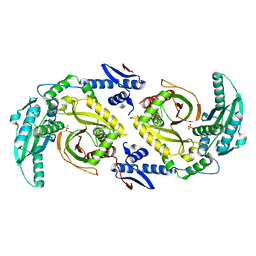 | | Molecular basis for condensation domain-mediated chain release from the enacyloxin polyketide synthase | | Descriptor: | Condensation domain protein, PHOSPHATE ION | | Authors: | Valentic, T.R, Tsai, S.C, Challis, G.L, Lewandowski, J.R, Kosol, S, Gallo, A, Griffiths, D, Masschelein, J.L, Jenner, M, De los Santos, E. | | Deposit date: | 2018-02-20 | | Release date: | 2019-02-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for chain release from the enacyloxin polyketide synthase.
Nat.Chem., 11, 2019
|
|
2K3J
 
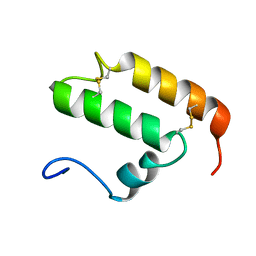 | | The solution structure of human Mia40 | | Descriptor: | Mitochondrial intermembrane space import and assembly protein 40 | | Authors: | Ciofi Baffoni, S, Bertini, I, Gallo, A. | | Deposit date: | 2008-05-08 | | Release date: | 2009-02-10 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | MIA40 is an oxidoreductase that catalyzes oxidative protein folding in mitochondria.
Nat.Struct.Mol.Biol., 16, 2009
|
|
2LGQ
 
 | | Human C30S/C59S-Cox17 mutant | | Descriptor: | Cytochrome c oxidase copper chaperone | | Authors: | Bertini, I, Ciofi-Baffoni, S, Gallo, A. | | Deposit date: | 2011-08-01 | | Release date: | 2011-08-17 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Functional role of two interhelical disulfide bonds in human cox17 protein from a structural perspective.
J.Biol.Chem., 286, 2011
|
|
2L0Y
 
 | | Complex hMia40-hCox17 | | Descriptor: | COX17 cytochrome c oxidase assembly homolog (S. cerevisiae) pseudogene (COX17), Mitochondrial intermembrane space import and assembly protein 40 | | Authors: | Bertini, I, Ciofi-Baffoni, S, Gallo, A. | | Deposit date: | 2010-07-20 | | Release date: | 2010-11-24 | | Last modified: | 2020-02-05 | | Method: | SOLUTION NMR | | Cite: | Molecular chaperone function of Mia40 triggers consecutive induced folding steps of the substrate in mitochondrial protein import.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
8RI9
 
 | | Late alpha-Synuclein fibril structure from liquid-liquid phase separations. | | Descriptor: | Alpha-synuclein | | Authors: | De Simone, A, Barritt, J.D, Chen, S, Cascella, R, Cecchi, C, Bigi, A, Jarvis, J.A, Chiti, F, Dobson, C.M, Fusco, G. | | Deposit date: | 2023-12-18 | | Release date: | 2024-03-06 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure-Toxicity Relationship in Intermediate Fibrils from alpha-Synuclein Condensates.
J.Am.Chem.Soc., 2024
|
|
8AQQ
 
 | |
8AQR
 
 | |
7P27
 
 | | NMR solution structure of Chikungunya virus macro domain | | Descriptor: | Polyprotein P1234 | | Authors: | Lykouras, M.V, Tsika, A.C, Papageorgiou, N, Canard, B, Coutard, B, Bentrop, D, Spyroulias, G.A. | | Deposit date: | 2021-07-04 | | Release date: | 2022-07-13 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Binding Adaptation of GS-441524 Diversifies Macro Domains and Downregulates SARS-CoV-2 de-MARylation Capacity.
J.Mol.Biol., 434, 2022
|
|
7QG7
 
 | | SARS-CoV-2 macrodomain Nsp3b bound to the remdesivir nucleoside GS-441524 | | Descriptor: | (2~{R},3~{R},4~{S},5~{R})-2-(4-azanylpyrrolo[2,1-f][1,2,4]triazin-7-yl)-5-(hydroxymethyl)-3,4-bis(oxidanyl)oxolane-2-carbonitrile, 1,2-ETHANEDIOL, Papain-like protease nsp3 | | Authors: | Wollenhaupt, J, Linhard, V, Sreeramulu, S, Weiss, M.S, Schwalbe, H. | | Deposit date: | 2021-12-07 | | Release date: | 2021-12-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Binding Adaptation of GS-441524 Diversifies Macro Domains and Downregulates SARS-CoV-2 de-MARylation Capacity.
J.Mol.Biol., 434, 2022
|
|
6F0C
 
 | |
6F0B
 
 | |
6TA8
 
 | |
