5K8A
 
 | | NIST FAB | | Descriptor: | Heavy chain, humanized Fab Lite chain | | Authors: | Gallagher, D.T, Galvin, C.V, Karageorgos, I, Marino, J.P. | | Deposit date: | 2016-05-27 | | Release date: | 2017-08-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Data on crystal organization in the structure of the Fab fragment from the NIST reference antibody, RM 8671.
Data Brief, 16, 2018
|
|
2FJT
 
 | | Adenylyl cyclase class iv from Yersinia pestis | | Descriptor: | Adenylyl cyclase class IV, SULFATE ION | | Authors: | Gallagher, D.T, Smith, N.N, Kim, S.-K, Reddy, P.T, Robinson, H, Heroux, A. | | Deposit date: | 2006-01-03 | | Release date: | 2006-11-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Structure of the class IV adenylyl cyclase reveals a novel fold
J.Mol.Biol., 362, 2006
|
|
5VGP
 
 | | Fc fragment of human IgG1 antibody, from NIST mAb | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Human Fc fragment with G1F/G0F glycan, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Gallagher, D.T, Galvin, C.V, Karageorgos, I, Marino, J.P. | | Deposit date: | 2017-04-11 | | Release date: | 2018-04-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.116 Å) | | Cite: | Structure of the Fc fragment of the NIST reference antibody RM8671.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
6P6D
 
 | | HUMAN IGG1 FC FRAGMENT, C239 INSERTION MUTANT | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CYSTEINE, IgG1 Fc fragment | | Authors: | Gallagher, D.T, Dimasi, N, McCullough, C. | | Deposit date: | 2019-06-03 | | Release date: | 2020-04-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structure and Dynamics of a Site-Specific Labeled Fc Fragment with Altered Effector Functions.
Pharmaceutics, 11, 2019
|
|
1OMO
 
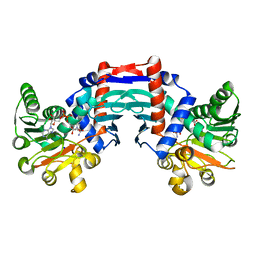 | | alanine dehydrogenase dimer w/bound NAD (archaeal) | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION, alanine dehydrogenase | | Authors: | Gallagher, D.T, Smith, N.N, Holden, M.J, Schroeder, I, Monbouquette, H.G. | | Deposit date: | 2003-02-25 | | Release date: | 2004-07-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structure of alanine dehydrogenase from Archaeoglobus: active site analysis and relation to bacterial cyclodeaminases and mammalian mu crystallin.
J.Mol.Biol., 342, 2004
|
|
1G1B
 
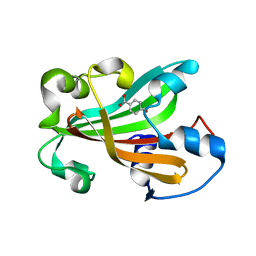 | | CHORISMATE LYASE (WILD-TYPE) WITH BOUND PRODUCT | | Descriptor: | CHORISMATE LYASE, P-HYDROXYBENZOIC ACID | | Authors: | Gallagher, D.T, Mayhew, M, Holden, M.J, Kim, K.J, Howard, A, Vilker, V.L. | | Deposit date: | 2000-10-11 | | Release date: | 2001-04-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The crystal structure of chorismate lyase shows a new fold and a tightly retained product.
Proteins, 44, 2001
|
|
1TDJ
 
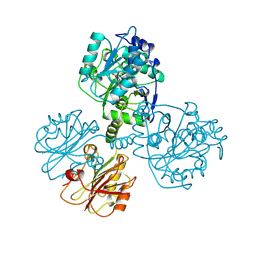 | | THREONINE DEAMINASE (BIOSYNTHETIC) FROM E. COLI | | Descriptor: | BIOSYNTHETIC THREONINE DEAMINASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Gallagher, D.T, Gilliland, G.L, Xiao, G, Eisenstein, E. | | Deposit date: | 1998-03-27 | | Release date: | 1998-10-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure and control of pyridoxal phosphate dependent allosteric threonine deaminase.
Structure, 6, 1998
|
|
3N0Y
 
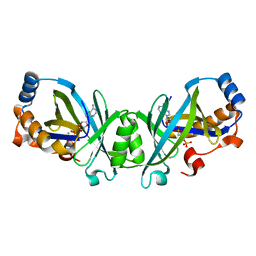 | | Adenylate cyclase class IV with active site ligand APC | | Descriptor: | Adenylate cyclase 2, DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, MANGANESE (II) ION | | Authors: | Gallagher, D.T, Reddy, P.T. | | Deposit date: | 2010-05-14 | | Release date: | 2010-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Active-Site Structure of Class IV Adenylyl Cyclase and Transphyletic Mechanism.
J.Mol.Biol., 405, 2011
|
|
3N0Z
 
 | |
3N10
 
 | | Product complex of adenylate cyclase class IV | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Adenylate cyclase 2, MANGANESE (II) ION, ... | | Authors: | Gallagher, D.T, Reddy, P.T. | | Deposit date: | 2010-05-14 | | Release date: | 2010-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Active-Site Structure of Class IV Adenylyl Cyclase and Transphyletic Mechanism.
J.Mol.Biol., 405, 2011
|
|
1FW9
 
 | | CHORISMATE LYASE WITH BOUND PRODUCT | | Descriptor: | CHORISMATE LYASE, P-HYDROXYBENZOIC ACID | | Authors: | Gallagher, D.T, Mayhew, M, Holden, M, Vilker, V, Howard, A. | | Deposit date: | 2000-09-22 | | Release date: | 2001-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The crystal structure of chorismate lyase shows a new fold and a tightly retained product.
Proteins, 44, 2001
|
|
1G81
 
 | | CHORISMATE LYASE WITH BOUND PRODUCT, ORTHORHOMBIC CRYSTAL FORM | | Descriptor: | CHORISMATE LYASE, P-HYDROXYBENZOIC ACID | | Authors: | Gallagher, D.T, Mayhew, M, Holden, M.J, Vilker, V.L, Howard, A. | | Deposit date: | 2000-11-15 | | Release date: | 2001-05-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | The crystal structure of chorismate lyase shows a new fold and a tightly retained product.
Proteins, 44, 2001
|
|
7RRW
 
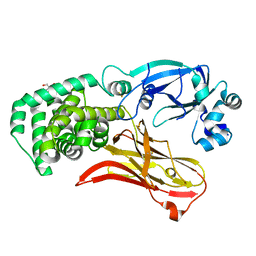 | | Monomeric CRM197 expressed in E. coli | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Diphtheria toxin | | Authors: | Gallagher, D.T, Lees, A. | | Deposit date: | 2021-08-10 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Monomeric crystal structure of the vaccine carrier protein CRM 197 and implications for vaccine development.
Acta Crystallogr.,Sect.F, 79, 2023
|
|
3D0S
 
 | |
1SUE
 
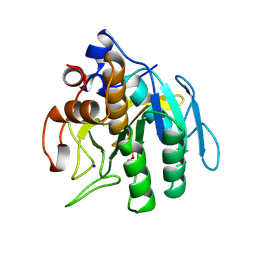 | | SUBTILISIN BPN' FROM BACILLUS AMYLOLIQUEFACIENS, MUTANT | | Descriptor: | DIISOPROPYL PHOSPHONATE, SODIUM ION, SUBTILISIN BPN' | | Authors: | Gallagher, D.T, Bryan, P, Pan, Q, Gilliland, G.L. | | Deposit date: | 1998-02-17 | | Release date: | 1998-10-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mechanism of ionic strength dependence of crystal growth rates in a subtilisin variant.
J.Cryst.Growth, 193, 1998
|
|
4ZRZ
 
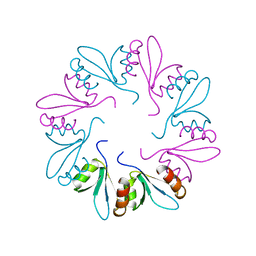 | | PlyCB mutant R66E | | Descriptor: | PlyCB | | Authors: | Gallagher, D.T, Nelson, D.C, Shen, Y. | | Deposit date: | 2015-05-12 | | Release date: | 2016-09-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | A bacteriophage endolysin that eliminates intracellular streptococci.
Elife, 5, 2016
|
|
8DNT
 
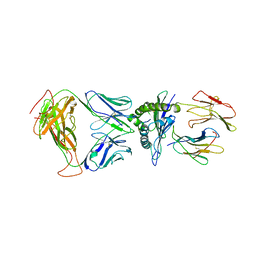 | | SARS-CoV-2 specific T cell receptor | | Descriptor: | Beta-2-microglobulin, MHC class I antigen alpha chain, Nucleoprotein, ... | | Authors: | Gallagher, D.T, Wu, D, Gowthaman, R, Pierce, B.G, Mariuzza, R.A, Weng, N.P. | | Deposit date: | 2022-07-11 | | Release date: | 2023-07-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | SARS-CoV-2 infection establishes a stable and age-independent CD8 + T cell response against a dominant nucleocapsid epitope using restricted T cell receptors.
Nat Commun, 14, 2023
|
|
2OX1
 
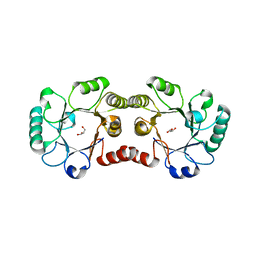 | | Archaeal Dehydroquinase | | Descriptor: | 3-dehydroquinate dehydratase, GLYCEROL | | Authors: | Gallagher, D.T, Smith, N.N. | | Deposit date: | 2007-02-19 | | Release date: | 2008-02-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structure and lability of archaeal dehydroquinase.
Acta Crystallogr.,Sect.F, 64, 2008
|
|
7KAE
 
 | |
2AHC
 
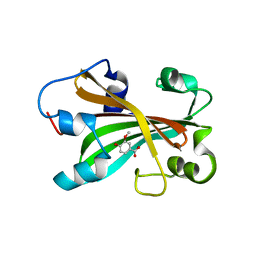 | | Chorismate lyase with inhibitor Vanilate | | Descriptor: | 4-HYDROXY-3-METHOXYBENZOATE, Chorismate lyase | | Authors: | Gallagher, D.T, Smith, N.N. | | Deposit date: | 2005-07-27 | | Release date: | 2006-01-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural analysis of ligand binding and catalysis in chorismate lyase
Arch.Biochem.Biophys., 445, 2006
|
|
7RHO
 
 | | Human IgG1 Fc fragment, hinge-free, expressed in E. coli | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Fc fragment of human IgG1 | | Authors: | Gallagher, D.T. | | Deposit date: | 2021-07-18 | | Release date: | 2022-11-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Effects of glycans and hinge on dynamics in the IgG1 Fc.
J.Biomol.Struct.Dyn., 2023
|
|
3CNQ
 
 | |
3F49
 
 | |
1TT8
 
 | | CHORISMATE LYASE WITH PRODUCT, 1.0 A RESOLUTION | | Descriptor: | Chorismate-pyruvate lyase, P-HYDROXYBENZOIC ACID | | Authors: | Gallagher, D.T, Mayhew, M, Holden, M.J, Vilker, V, Howard, A. | | Deposit date: | 2004-06-22 | | Release date: | 2004-12-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Structural analysis of ligand binding and catalysis in chorismate lyase.
Arch.Biochem.Biophys., 445, 2006
|
|
1XLR
 
 | | CHORISMATE LYASE WITH INHIBITOR VANILLATE | | Descriptor: | 4-HYDROXY-3-METHOXYBENZOATE, Chorismate--pyruvate lyase | | Authors: | Gallagher, D.T, Smith, N. | | Deposit date: | 2004-09-30 | | Release date: | 2004-11-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural analysis of ligand binding and catalysis in chorismate lyase.
Arch.Biochem.Biophys., 445, 2006
|
|
