7OWD
 
 | |
7OWC
 
 | |
4OYK
 
 | |
3PV1
 
 | | Crystal structure of the USP15 DUSP-UBL domains | | Descriptor: | ACETATE ION, Ubiquitin carboxyl-terminal hydrolase 15 | | Authors: | Elliott, P.R, Urbe, S, Clague, M.J, Barsukov, I.L. | | Deposit date: | 2010-12-06 | | Release date: | 2011-11-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural variability of the ubiquitin specific protease DUSP-UBL double domains.
Febs Lett., 585, 2011
|
|
4OYJ
 
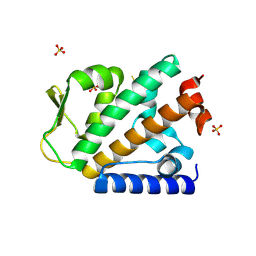 | | Structure of the apo HOIP PUB domain | | Descriptor: | E3 ubiquitin-protein ligase RNF31, SULFATE ION | | Authors: | Elliott, P.R, Komander, D. | | Deposit date: | 2014-02-12 | | Release date: | 2014-05-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Molecular Basis and Regulation of OTULIN-LUBAC Interaction.
Mol.Cell, 54, 2014
|
|
3IVF
 
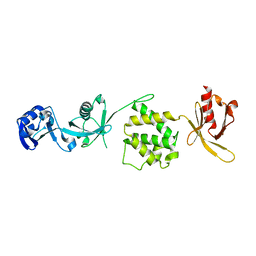 | | Crystal structure of the talin head FERM domain | | Descriptor: | Talin-1 | | Authors: | Elliott, P.R, Goult, B.T, Bate, N, Grossmann, J.G, Roberts, G.C.K, Critchley, D.R, Barsukov, I.L. | | Deposit date: | 2009-09-01 | | Release date: | 2010-08-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | The Structure of the talin head reveals a novel extended conformation of the FERM domain
Structure, 18, 2010
|
|
5LJM
 
 | | Structure of SPATA2 PUB domain | | Descriptor: | GLYCEROL, Spermatogenesis-associated protein 2 | | Authors: | Elliott, P.R, Komander, D. | | Deposit date: | 2016-07-18 | | Release date: | 2016-08-24 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.454 Å) | | Cite: | SPATA2 Links CYLD to LUBAC, Activates CYLD, and Controls LUBAC Signaling.
Mol.Cell, 63, 2016
|
|
5LJN
 
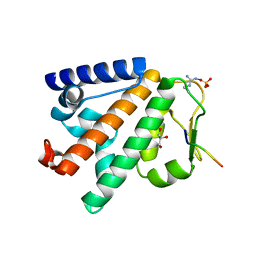 | | Structure of the HOIP PUB domain bound to SPATA2 PIM peptide | | Descriptor: | E3 ubiquitin-protein ligase RNF31, GLYCEROL, SULFATE ION, ... | | Authors: | Elliott, P.R, Komander, D. | | Deposit date: | 2016-07-18 | | Release date: | 2016-08-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | SPATA2 Links CYLD to LUBAC, Activates CYLD, and Controls LUBAC Signaling.
Mol.Cell, 63, 2016
|
|
4A3P
 
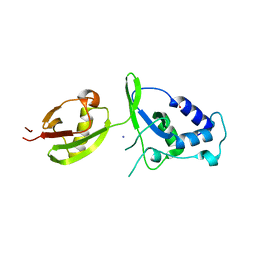 | | Structure of USP15 DUSP-UBL deletion mutant | | Descriptor: | ACETATE ION, IODIDE ION, UBIQUITIN CARBOXYL-TERMINAL HYDROLASE 15 | | Authors: | Elliott, P.R, Liu, H, Pastok, M.W, Grossmann, G.J, Rigden, D.J, Clague, M.J, Urbe, S, Barsukov, I.L. | | Deposit date: | 2011-10-03 | | Release date: | 2011-11-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural Variability of the Ubiquitin Specific Protease Dusp-Ubl Double Domains.
FEBS Lett., 585, 2011
|
|
4A3O
 
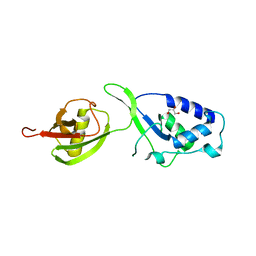 | | Crystal structure of the USP15 DUSP-UBL monomer | | Descriptor: | GLYCEROL, UBIQUITIN CARBOXYL-TERMINAL HYDROLASE 15 | | Authors: | Elliott, P.R, Liu, H, Pastok, M.W, Grossmann, G.J, Rigden, D.J, Clague, M.J, Urbe, S, Barsukov, I.L. | | Deposit date: | 2011-10-03 | | Release date: | 2011-11-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Variability of the Ubiquitin Specific Protease Dusp-Ubl Double Domains.
FEBS Lett., 585, 2011
|
|
1QLP
 
 | | 2.0 ANGSTROM STRUCTURE OF INTACT ALPHA-1-ANTITRYPSIN: A CANONICAL TEMPLATE FOR ACTIVE SERPINS | | Descriptor: | ALPHA-1-ANTITRYPSIN | | Authors: | Elliott, P.R, Pei, X.Y, Dafforn, T, Read, R.J, Carrell, R.W, Lomas, D.A. | | Deposit date: | 1999-09-10 | | Release date: | 1999-09-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Topography of a 2.0 A structure of alpha1-antitrypsin reveals targets for rational drug design to prevent conformational disease.
Protein Sci., 9, 2000
|
|
6SAK
 
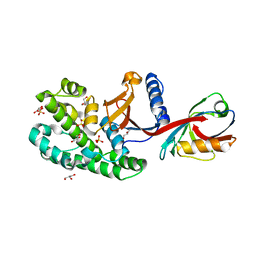 | |
5OE7
 
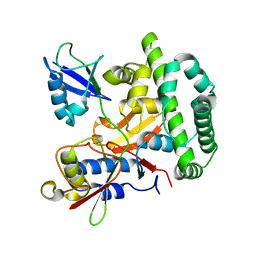 | |
2LNK
 
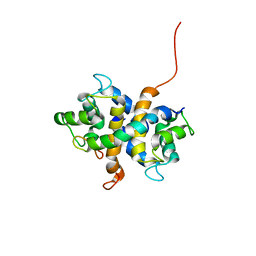 | |
1PSI
 
 | |
6I9C
 
 | | Structure of the OTU domain of OTULIN G281R mutant | | Descriptor: | CHLORIDE ION, GLYCEROL, Ubiquitin thioesterase otulin | | Authors: | Damgaard, R.B, Elliott, P.R, Swatek, K.N, Maher, E.R, Stepensky, P, Elpeleg, O, Komander, D, Berkun, Y. | | Deposit date: | 2018-11-22 | | Release date: | 2019-03-06 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | OTULIN deficiency in ORAS causes cell type-specific LUBAC degradation, dysregulated TNF signalling and cell death.
Embo Mol Med, 11, 2019
|
|
8ATM
 
 | |
8ATO
 
 | |
8P5P
 
 | | Structure of TECPR1 N-terminal DysF domain | | Descriptor: | GLYCEROL, Tectonin beta-propeller repeat-containing protein 1 | | Authors: | Boyle, K.B, Elliott, P.R, Randow, F. | | Deposit date: | 2023-05-24 | | Release date: | 2023-07-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | TECPR1 conjugates LC3 to damaged endomembranes upon detection of sphingomyelin exposure.
Embo J., 42, 2023
|
|
6YIE
 
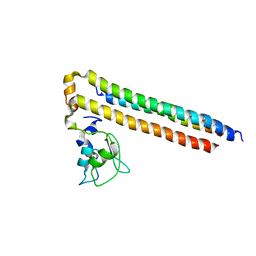 | | Structure of a Borealin-INCENP-Survivin complex | | Descriptor: | Baculoviral IAP repeat-containing protein 5, Borealin, Inner centromere protein, ... | | Authors: | Serena, M, Elliott, P.R, Barr, F.A. | | Deposit date: | 2020-04-01 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.49 Å) | | Cite: | Molecular basis of MKLP2-dependent Aurora B transport from chromatin to the anaphase central spindle.
J.Cell Biol., 219, 2020
|
|
6YIF
 
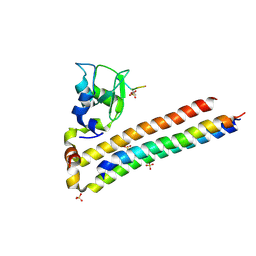 | | Structure of Chromosomal Passenger Complex (CPC) bound to phosphorylated Histone 3 peptide at 1.8 A. | | Descriptor: | Baculoviral IAP repeat-containing protein 5, Borealin, Inner centromere protein, ... | | Authors: | Serena, M, Elliott, P.R, Barr, F.A. | | Deposit date: | 2020-04-01 | | Release date: | 2020-05-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Molecular basis of MKLP2-dependent Aurora B transport from chromatin to the anaphase central spindle.
J.Cell Biol., 219, 2020
|
|
6YIH
 
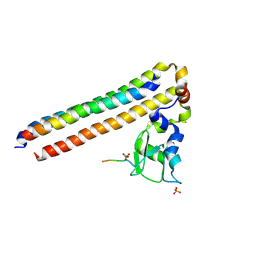 | | Structure of Chromosomal Passenger Complex (CPC) bound to phosphorylated Histone 3 peptide at 2.6 A. | | Descriptor: | Baculoviral IAP repeat-containing protein 5, Borealin, Histone H3.1, ... | | Authors: | Serena, M, Elliott, P.R, Barr, F.A. | | Deposit date: | 2020-04-01 | | Release date: | 2020-05-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Molecular basis of MKLP2-dependent Aurora B transport from chromatin to the anaphase central spindle.
J.Cell Biol., 219, 2020
|
|
6YIP
 
 | | Structure of MKLP2 coiled coil | | Descriptor: | ISOPROPYL ALCOHOL, Kinesin-like protein KIF20A | | Authors: | Serena, M, Elliott, P.R, Barr, F.A. | | Deposit date: | 2020-04-01 | | Release date: | 2020-05-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Molecular basis of MKLP2-dependent Aurora B transport from chromatin to the anaphase central spindle.
J.Cell Biol., 219, 2020
|
|
5LRU
 
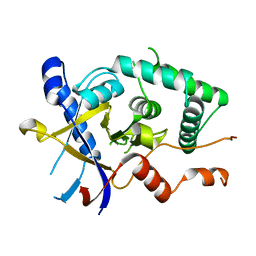 | | Structure of Cezanne/OTUD7B OTU domain | | Descriptor: | OTU domain-containing protein 7B | | Authors: | Mevissen, T.E.T, Kulathu, Y, Mulder, M.P.C, Geurink, P.P, Maslen, S.L, Gersch, M, Elliott, P.R, Burke, J.E, van Tol, B.D.M, Akutsu, M, El Oualid, F, Kawasaki, M, Freund, S.M.V, Ovaa, H, Komander, D. | | Deposit date: | 2016-08-22 | | Release date: | 2016-10-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular basis of Lys11-polyubiquitin specificity in the deubiquitinase Cezanne.
Nature, 538, 2016
|
|
5LRX
 
 | | Structure of A20 OTU domain bound to ubiquitin | | Descriptor: | Polyubiquitin-B, Tumor necrosis factor alpha-induced protein 3 | | Authors: | Mevissen, T.E.T, Kulathu, Y, Mulder, M.P.C, Geurink, P.P, Maslen, S.L, Gersch, M, Elliott, P.R, Burke, J.E, van Tol, B.D.M, Akutsu, M, El Oualid, F, Kawasaki, M, Freund, S.M.V, Ovaa, H, Komander, D. | | Deposit date: | 2016-08-22 | | Release date: | 2016-10-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Molecular basis of Lys11-polyubiquitin specificity in the deubiquitinase Cezanne.
Nature, 538, 2016
|
|
