3K4T
 
 | |
3H0I
 
 | |
3H0H
 
 | |
1NDK
 
 | | X-RAY STRUCTURE OF NUCLEOSIDE DIPHOSPHATE KINASE | | Descriptor: | NUCLEOSIDE DIPHOSPHATE KINASE | | Authors: | Janin, J, Dumas, C, Morera, S, Lascu, I, Veron, M. | | Deposit date: | 1993-07-15 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray structure of nucleoside diphosphate kinase.
EMBO J., 11, 1992
|
|
1AVV
 
 | | HIV-1 NEF PROTEIN, UNLIGANDED CORE DOMAIN | | Descriptor: | NEGATIVE FACTOR | | Authors: | Arold, S, Franken, P, Dumas, C. | | Deposit date: | 1997-09-21 | | Release date: | 1998-03-25 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The crystal structure of HIV-1 Nef protein bound to the Fyn kinase SH3 domain suggests a role for this complex in altered T cell receptor signaling.
Structure, 5, 1997
|
|
1AVZ
 
 | |
1BU1
 
 | | SRC FAMILY KINASE HCK SH3 DOMAIN | | Descriptor: | PROTEIN (HEMOPOIETIC CELL KINASE) | | Authors: | Arold, S, Franken, P, Dumas, C. | | Deposit date: | 1998-09-09 | | Release date: | 1998-11-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | RT loop flexibility enhances the specificity of Src family SH3 domains for HIV-1 Nef.
Biochemistry, 37, 1998
|
|
4QFT
 
 | |
5HT6
 
 | | Crystal structure of the SET domain of the human MLL5 methyltransferase | | Descriptor: | Histone-lysine N-methyltransferase 2E | | Authors: | le Maire, A, Mas-y-Mas, S, Dumas, C, Lebedev, A. | | Deposit date: | 2016-01-26 | | Release date: | 2016-11-16 | | Method: | X-RAY DIFFRACTION (2.093 Å) | | Cite: | The Human Mixed Lineage Leukemia 5 (MLL5), a Sequentially and Structurally Divergent SET Domain-Containing Protein with No Intrinsic Catalytic Activity.
Plos One, 11, 2016
|
|
4F7O
 
 | | Crystal structure of CSN5 | | Descriptor: | COP9 signalosome complex subunit 5, THIOCYANATE ION, ZINC ION | | Authors: | Echalier, A, Birol, M, Hoh, F, Dumas, C. | | Deposit date: | 2012-05-16 | | Release date: | 2013-01-23 | | Last modified: | 2013-02-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Insights into the regulation of the human COP9 signalosome catalytic subunit, CSN5/Jab1.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
1NDP
 
 | | ADENOSINE 5'-DIPHOSPHATE BINDING AND THE ACTIVE SITE OF NUCLEOSIDE DIPHOSPHATE KINASE | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, NUCLEOSIDE DIPHOSPHATE KINASE | | Authors: | Janin, J, Morera, S, Dumas, C, Lascu, I, Lebras, G, Veron, M. | | Deposit date: | 1993-11-29 | | Release date: | 1994-04-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Adenosine 5'-diphosphate binding and the active site of nucleoside diphosphate kinase.
Biochemistry, 33, 1994
|
|
1NDL
 
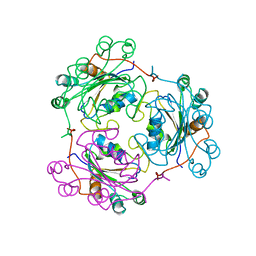 | | THE AWD NUCLEOTIDE DIPHOSPHATE KINASE FROM DROSOPHILA | | Descriptor: | NUCLEOSIDE DIPHOSPHATE KINASE | | Authors: | Janin, J, Chiadmi, M, Dumas, C, Lascu, I, Lebras, G, Morera, S, Veron, M. | | Deposit date: | 1993-11-27 | | Release date: | 1994-04-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the Awd nucleotide diphosphate kinase from Drosophila.
Structure, 1, 1993
|
|
3H0F
 
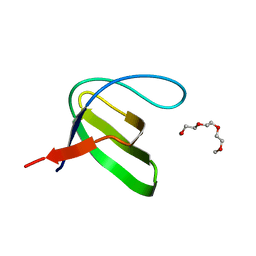 | | Crystal structure of the human Fyn SH3 R96W mutant | | Descriptor: | 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, Proto-oncogene tyrosine-protein kinase Fyn | | Authors: | Ponchon, L, Hoh, F, Labesse, G, Dumas, C, Arold, S.T. | | Deposit date: | 2009-04-09 | | Release date: | 2010-04-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Synergy and allostery in ligand binding by HIV-1 Nef.
Biochem.J., 478, 2021
|
|
4D8D
 
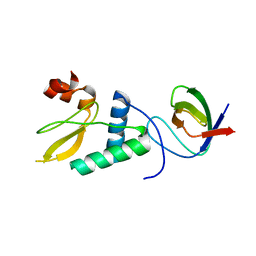 | |
1G33
 
 | | CRYSTAL STRUCTURE OF RAT PARVALBUMIN WITHOUT THE N-TERMINAL DOMAIN | | Descriptor: | CALCIUM ION, PARVALBUMIN ALPHA, SULFATE ION | | Authors: | Thepaut, M, Strub, M.P, Cave, A, Baneres, J.L, Berchtold, M.W, Dumas, C, Padilla, A. | | Deposit date: | 2000-10-23 | | Release date: | 2001-10-03 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Structure of rat parvalbumin with deleted AB domain: implications for the evolution of EF hand calcium-binding proteins and possible physiological relevance.
Proteins, 45, 2001
|
|
3F6N
 
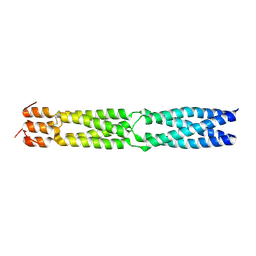 | |
1PAE
 
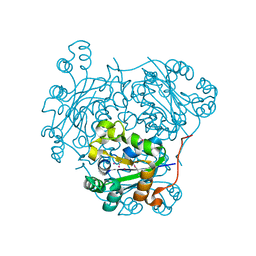 | | nucleoside diphosphate kinase | | Descriptor: | Nucleoside diphosphate kinase, cytosolic, SELENIUM ATOM | | Authors: | Strub, M.-P, Hoh, F, Sanchez, J.-F, Strub, J.M, Bock, A, Aumelas, A, Dumas, C. | | Deposit date: | 2003-05-14 | | Release date: | 2003-11-11 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Selenomethionine and Selenocysteine Double Labeling Strategy for Crystallographic Phasing
Structure, 11, 2003
|
|
1PFP
 
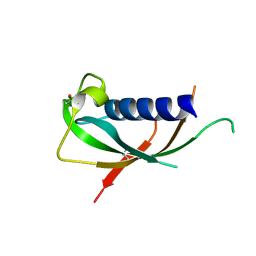 | | CATHELIN-LIKE MOTIF OF PROTEGRIN-3 | | Descriptor: | Protegrin 3 | | Authors: | Strub, M.-P, Hoh, F, Sanchez, J.-F, Strub, J.M, Bock, A, Aumelas, A, Dumas, C. | | Deposit date: | 2003-05-27 | | Release date: | 2003-11-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Selenomethionine and Selenocysteine Double Labeling Strategy for Crystallographic Phasing
Structure, 11, 2003
|
|
1KWI
 
 | | Crystal Structure Analysis of the Cathelicidin Motif of Protegrins | | Descriptor: | Protegrin-3 Precursor | | Authors: | Sanchez, J.F, Hoh, F, Strub, M.P, Aumelas, A, Dumas, C. | | Deposit date: | 2002-01-29 | | Release date: | 2002-10-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structure of the cathelicidin motif of protegrin-3 precursor: structural insights into the activation mechanism of an antimicrobial protein.
Structure, 10, 2002
|
|
1LXE
 
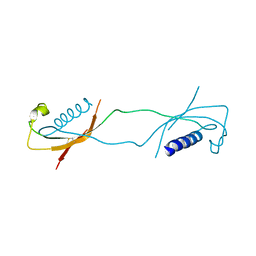 | | CRYSTAL STRUCTURE OF THE CATHELICIDIN MOTIF OF PROTEGRINS | | Descriptor: | protegrin-3 precursor | | Authors: | Sanchez, J.F, Hoh, F, Strub, M.P, Aumelas, A, Dumas, C. | | Deposit date: | 2002-06-05 | | Release date: | 2002-10-09 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the cathelicidin motif of protegrin-3 precursor: structural insights into the activation mechanism of an antimicrobial protein.
Structure, 10, 2002
|
|
1R3B
 
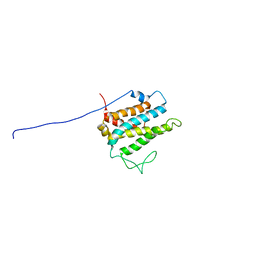 | | Solution structure of xenopus laevis Mob1 | | Descriptor: | MOB1 | | Authors: | Ponchon, L, Dumas, C, Kajava, A.V, Fesquet, D, Padilla, A. | | Deposit date: | 2003-10-01 | | Release date: | 2004-09-28 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of Mob1, a mitotic exit network protein and its interaction with an NDR kinase peptide
J.Mol.Biol., 337, 2004
|
|
1T7H
 
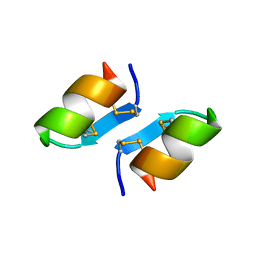 | | X-ray structure of [Lys(-2)-Arg(-1)-des(17-21)]-endothelin-1 peptide | | Descriptor: | Endothelin-1 | | Authors: | Hoh, F, Cerdan, R, Kaas, Q, Nishi, Y, Chiche, L, Kubo, S, Chino, N, Kobayashi, Y, Dumas, C, Aumelas, A. | | Deposit date: | 2004-05-10 | | Release date: | 2004-12-21 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | High-resolution X-ray structure of the unexpectedly stable dimer of the [Lys(-2)-Arg(-1)-des(17-21)]endothelin-1 peptide
Biochemistry, 43, 2004
|
|
1T6F
 
 | | Crystal Structure of the Coiled-coil Dimerization Motif of Geminin | | Descriptor: | Geminin | | Authors: | Thepaut, M, Maiorano, D, Guichou, J.-F, Auge, M.-T, Dumas, C, Mechali, M, Padilla, A. | | Deposit date: | 2004-05-06 | | Release date: | 2004-07-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Crystal Structure of the Coiled-coil Dimerization Motif of Geminin: Structural and Functional Insights on DNA Replication Regulation
J.Mol.Biol., 342, 2004
|
|
1TUK
 
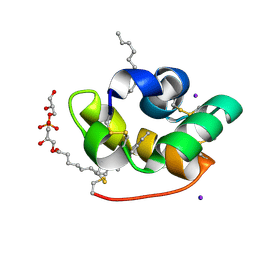 | | Crystal structure of liganded type 2 non specific lipid transfer protein from wheat | | Descriptor: | 1-MYRISTOYL-2-HYDROXY-SN-GLYCERO-3-[PHOSPHO-RAC-(1-GLYCEROL)], IODIDE ION, Nonspecific lipid-transfer protein 2G | | Authors: | Hoh, F, Pons, J.L, Gautier, M.F, De Lamotte, F, Dumas, C. | | Deposit date: | 2004-06-25 | | Release date: | 2005-04-05 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Structure of a liganded type 2 non-specific lipid-transfer protein from wheat and the molecular basis of lipid binding.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
7D7S
 
 | |
