1JW4
 
 | | Structure of ligand-free maltodextrin-binding protein | | Descriptor: | maltodextrin-binding protein | | Authors: | Duan, X, Quiocho, F.A. | | Deposit date: | 2001-09-02 | | Release date: | 2002-01-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural evidence for a dominant role of nonpolar interactions in the binding of a transport/chemosensory receptor to its highly polar ligands.
Biochemistry, 41, 2002
|
|
1JW5
 
 | |
3WNV
 
 | | Crystal structure of a glyoxylate reductase from Paecilomyes thermophila | | Descriptor: | SULFATE ION, glyoxylate reductase | | Authors: | Duan, X, Hu, S, Zhou, P, Zhou, Y, Jiang, Z. | | Deposit date: | 2013-12-17 | | Release date: | 2014-12-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Characterization and crystal structure of a first fungal glyoxylate reductase from Paecilomyes thermophila
Enzyme.Microb.Technol., 60, 2014
|
|
1VDE
 
 | |
3HTK
 
 | | Crystal structure of Mms21 and Smc5 complex | | Descriptor: | E3 SUMO-protein ligase MMS21, Structural maintenance of chromosomes protein 5, ZINC ION | | Authors: | Duan, X, Sarangi, P, Liu, X, Rangi, G.K, Zhao, X, Ye, H. | | Deposit date: | 2009-06-11 | | Release date: | 2009-10-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structural and functional insights into the roles of the Mms21 subunit of the Smc5/6 complex.
Mol.Cell, 35, 2009
|
|
1FQA
 
 | | STRUCTURE OF MALTOTETRAITOL BOUND TO OPEN-FORM MALTODEXTRIN BINDING PROTEIN IN P2(1)CRYSTAL FORM | | Descriptor: | MALTODEXTRIN-BINDING PROTEIN, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-sorbitol | | Authors: | Duan, X, Hall, J.A, Nikaido, H, Quiocho, F.A. | | Deposit date: | 2000-09-04 | | Release date: | 2001-03-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of the maltodextrin/maltose-binding protein complexed with reduced oligosaccharides: flexibility of tertiary structure and ligand binding.
J.Mol.Biol., 306, 2001
|
|
1FQC
 
 | | CRYSTAL STRUCTURE OF MALTOTRIOTOL BOUND TO CLOSED-FORM MALTODEXTRIN BINDING PROTEIN | | Descriptor: | MALTODEXTRIN-BINDING PROTEIN, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-D-glucose | | Authors: | Duan, X, Hall, J.A, Nikaido, H, Quiocho, F.A. | | Deposit date: | 2000-09-04 | | Release date: | 2001-03-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of the maltodextrin/maltose-binding protein complexed with reduced oligosaccharides: flexibility of tertiary structure and ligand binding.
J.Mol.Biol., 306, 2001
|
|
1FQD
 
 | | CRYSTAL STRUCTURE OF MALTOTETRAITOL BOUND TO CLOSED-FORM MALTODEXTRIN BINDING PROTEIN | | Descriptor: | MALTODEXTRIN-BINDING PROTEIN, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-D-glucose | | Authors: | Duan, X, Hall, J.A, Nikaido, H, Quiocho, F.A. | | Deposit date: | 2000-09-04 | | Release date: | 2001-03-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of the maltodextrin/maltose-binding protein complexed with reduced oligosaccharides: flexibility of tertiary structure and ligand binding.
J.Mol.Biol., 306, 2001
|
|
1FQB
 
 | | STRUCTURE OF MALTOTRIOTOL BOUND TO OPEN-FORM MALTODEXTRIN BINDING PROTEIN IN P2(1)CRYSTAL FORM | | Descriptor: | MALTODEXTRIN-BINDING PROTEIN, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-sorbitol | | Authors: | Duan, X, Hall, J.A, Nikaido, H, Quiocho, F.A. | | Deposit date: | 2000-09-04 | | Release date: | 2001-03-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of the maltodextrin/maltose-binding protein complexed with reduced oligosaccharides: flexibility of tertiary structure and ligand binding.
J.Mol.Biol., 306, 2001
|
|
1EZ9
 
 | |
4WY5
 
 | | Structural analysis of two fungal esterases from Rhizomucor miehei explaining their substrate specificity | | Descriptor: | Esterase, SULFATE ION | | Authors: | Qin, Z, Yang, S, Duan, X, Yan, Q, Jiang, Z. | | Deposit date: | 2014-11-15 | | Release date: | 2015-07-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structural insights into the substrate specificity of two esterases from the thermophilic Rhizomucor miehei
J.Lipid Res., 56, 2015
|
|
4WY8
 
 | | Structural analysis of two fungal esterases from Rhizomucor miehei explaining their substrate specificity | | Descriptor: | esterase | | Authors: | Qin, Z, Yang, S, Duan, X, Yan, Q, Jiang, Z. | | Deposit date: | 2014-11-16 | | Release date: | 2015-07-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structural insights into the substrate specificity of two esterases from the thermophilic Rhizomucor miehei
J.Lipid Res., 56, 2015
|
|
8J6I
 
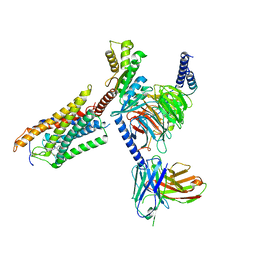 | | Cryo-EM structure of thehydroxycarboxylic acid receptor 2-Gi protein complex bound MK-6892 | | Descriptor: | 2-[[2,2-dimethyl-3-[3-(5-oxidanylpyridin-2-yl)-1,2,4-oxadiazol-5-yl]propanoyl]amino]cyclohexene-1-carboxylic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Yuan, Q, Zhu, S, Duan, J, Xu, H.E, Duan, X. | | Deposit date: | 2023-04-26 | | Release date: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Molecular recognition of niacin and lipid-lowering drugs by the human hydroxycarboxylic acid receptor 2.
Cell Rep, 42, 2023
|
|
8J6L
 
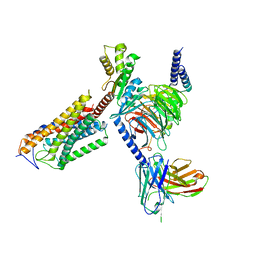 | | Cryo-EM structure of thehydroxycarboxylic acid receptor 2-Gi protein complex bound niacin | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Yuan, Q, Zhu, S, Duan, J, Xu, H.E, Duan, X. | | Deposit date: | 2023-04-26 | | Release date: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Molecular recognition of niacin and lipid-lowering drugs by the human hydroxycarboxylic acid receptor 2.
Cell Rep, 42, 2023
|
|
4NXL
 
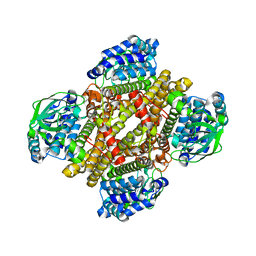 | | Dibenzothiophene monooxygenase (DszC) from Rhodococcus erythropolis | | Descriptor: | DszC | | Authors: | Zhang, L, Duan, X, Li, X, Rao, Z. | | Deposit date: | 2013-12-09 | | Release date: | 2014-07-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insights into the stabilization of active, tetrameric DszC by its C-terminus.
Proteins, 82, 2014
|
|
1DFA
 
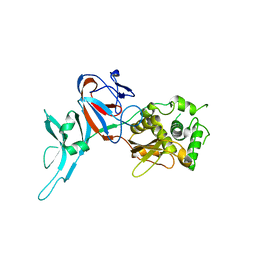 | | CRYSTAL STRUCTURE OF PI-SCEI IN C2 SPACE GROUP | | Descriptor: | PI-SCEI ENDONUCLEASE | | Authors: | Hu, D, Crist, M, Duan, X, Quiocho, F.A, Gimble, F.S. | | Deposit date: | 1999-11-18 | | Release date: | 1999-12-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Probing the structure of the PI-SceI-DNA complex by affinity cleavage and affinity photocross-linking.
J.Biol.Chem., 275, 2000
|
|
1DVP
 
 | | CRYSTAL STRUCTURE OF THE VHS AND FYVE TANDEM DOMAINS OF HRS, A PROTEIN INVOLVED IN MEMBRANE TRAFFICKING AND SIGNAL TRANSDUCTION | | Descriptor: | CITRIC ACID, HEPATOCYTE GROWTH FACTOR-REGULATED TYROSINE KINASE SUBSTRATE, ZINC ION | | Authors: | Mao, Y, Nickitenko, A, Duan, X, Lloyd, T.E, Wu, M.N, Bellen, H, Quiocho, F.A. | | Deposit date: | 2000-01-21 | | Release date: | 2000-03-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the VHS and FYVE tandem domains of Hrs, a protein involved in membrane trafficking and signal transduction.
Cell(Cambridge,Mass.), 100, 2000
|
|
8J6J
 
 | | Cryo-EM structure of thehydroxycarboxylic acid receptor 2-Gi protein complex bound with GSK256073 | | Descriptor: | 8-chloranyl-3-pentyl-7H-purine-2,6-dione, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Yuan, Q, Zhu, S, Duan, J, Xu, H.E, Duan, X. | | Deposit date: | 2023-04-26 | | Release date: | 2024-01-10 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Molecular recognition of niacin and lipid-lowering drugs by the human hydroxycarboxylic acid receptor 2.
Cell Rep, 42, 2023
|
|
4JEK
 
 | | Structure of dibenzothiophene monooxygenase (DszC) from Rhodococcus erythropolis | | Descriptor: | Dibenzothiophene desulfurization enzyme C | | Authors: | Zhang, L, Duan, X.L, Zhou, D.M, Li, X. | | Deposit date: | 2013-02-27 | | Release date: | 2013-09-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystallization and preliminary structural analysis of dibenzothiophene monooxygenase (DszC) from Rhodococcus erythropolis
Acta Crystallogr.,Sect.F, 69, 2013
|
|
5X88
 
 | | A crystal structure of cutinases from Malbranchea cinnamomea | | Descriptor: | cutinase | | Authors: | Jiang, Z, Yang, S.Q, You, X, Huang, P, Ma, J.W. | | Deposit date: | 2017-03-01 | | Release date: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | High-level expression and characterization of a novel cutinase from Malbranchea cinnamomea suitable for butyl butyrate production.
Biotechnol Biofuels, 10, 2017
|
|
7WB5
 
 | | local structure of hu33 and spike | | Descriptor: | Surface glycoprotein, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, hu33 heavy chain, ... | | Authors: | Pulan, L. | | Deposit date: | 2021-12-15 | | Release date: | 2022-10-26 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | A non-ACE2-blocking neutralizing antibody against Omicron-included SARS-CoV-2 variants.
Signal Transduct Target Ther, 7, 2022
|
|
7WBH
 
 | | overall structure of hu33 and spike | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Pulan, L. | | Deposit date: | 2021-12-16 | | Release date: | 2022-10-26 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | A non-ACE2-blocking neutralizing antibody against Omicron-included SARS-CoV-2 variants.
Signal Transduct Target Ther, 7, 2022
|
|
1VFQ
 
 | |
3VHL
 
 | | Crystal structure of the DHR-2 domain of DOCK8 in complex with Cdc42 (T17N mutant) | | Descriptor: | Cell division control protein 42 homolog, Dedicator of cytokinesis protein 8, PHOSPHATE ION | | Authors: | Hanawa-Suetsugu, K, Kukimoto-Niino, M, Nishizak, T, Terada, T, Shirouzu, M, Fukui, Y, Yokoyama, S. | | Deposit date: | 2011-08-26 | | Release date: | 2012-06-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.085 Å) | | Cite: | DOCK8 is a Cdc42 activator critical for interstitial dendritic cell migration during immune responses.
Blood, 119, 2012
|
|
1LWS
 
 | | Crystal structure of the intein homing endonuclease PI-SceI bound to its recognition sequence | | Descriptor: | CALCIUM ION, ENDONUCLEASE PI-SCEI, PI-SceI DNA recognition region bottom strand, ... | | Authors: | Moure, C.M, Gimble, F.S, Quiocho, F.A. | | Deposit date: | 2002-06-03 | | Release date: | 2002-09-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structure of the intein homing endonuclease PI-SceI bound to its recognition sequence.
Nat.Struct.Biol., 9, 2002
|
|
