1BMT
 
 | | HOW A PROTEIN BINDS B12: A 3.O ANGSTROM X-RAY STRUCTURE OF THE B12-BINDING DOMAINS OF METHIONINE SYNTHASE | | Descriptor: | CO-METHYLCOBALAMIN, METHIONINE SYNTHASE | | Authors: | Drennan, C.L, Huang, S, Drummond, J.T, Matthews, R.G, Ludwig, M.L. | | Deposit date: | 1994-09-02 | | Release date: | 1995-06-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | How a protein binds B12: A 3.0 A X-ray structure of B12-binding domains of methionine synthase.
Science, 266, 1994
|
|
1JQK
 
 | | Crystal structure of carbon monoxide dehydrogenase from Rhodospirillum rubrum | | Descriptor: | FE (II) ION, FE(3)-NI(1)-S(4) CLUSTER, IRON/SULFUR CLUSTER, ... | | Authors: | Drennan, C.L, Heo, J, Sintchak, M.D, Schreiter, E, Ludden, P.W. | | Deposit date: | 2001-08-07 | | Release date: | 2001-10-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Life on carbon monoxide: X-ray structure of Rhodospirillum rubrum Ni-Fe-S carbon monoxide dehydrogenase.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1D03
 
 | | REFINED STRUCTURES OF OXIDIZED FLAVODOXIN FROM ANACYSTIS NIDULANS | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVODOXIN | | Authors: | Drennan, C.L, Pattridge, K.A, Weber, C.H, Metzger, A.L, Hoover, D.M, Ludwig, M.L. | | Deposit date: | 1999-09-08 | | Release date: | 1999-12-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Refined structures of oxidized flavodoxin from Anacystis nidulans.
J.Mol.Biol., 294, 1999
|
|
1CZU
 
 | | REFINED STRUCTURES OF OXIDIZED FLAVODOXIN FROM ANACYSTIS NIDULANS | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVODOXIN | | Authors: | Drennan, C.L, Pattridge, K.A, Weber, C.H, Metzger, A.L, Hoover, D.M, Ludwig, M.L. | | Deposit date: | 1999-09-07 | | Release date: | 1999-12-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Refined structures of oxidized flavodoxin from Anacystis nidulans.
J.Mol.Biol., 294, 1999
|
|
4XJK
 
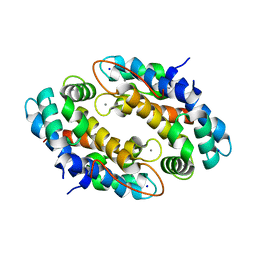 | | Crystal structure of Mn(II) Ca(II) Na(I) bound calprotectin | | Descriptor: | CALCIUM ION, MANGANESE (II) ION, Protein S100-A8, ... | | Authors: | Drennan, C.L, Bowman, S.E.J. | | Deposit date: | 2015-01-08 | | Release date: | 2015-04-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Manganese Binding Properties of Human Calprotectin under Conditions of High and Low Calcium: X-ray Crystallographic and Advanced Electron Paramagnetic Resonance Spectroscopic Analysis.
J.Am.Chem.Soc., 137, 2015
|
|
3SCH
 
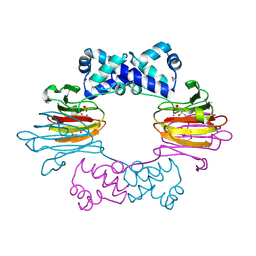 | | Co(II)-HppE with R-HPP | | Descriptor: | COBALT (II) ION, Epoxidase, [(2R)-2-hydroxypropyl]phosphonic acid | | Authors: | Drennan, C.L. | | Deposit date: | 2011-06-07 | | Release date: | 2011-07-06 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of regiospecificity of a mononuclear iron enzyme in antibiotic fosfomycin biosynthesis.
J.Am.Chem.Soc., 133, 2011
|
|
3SCG
 
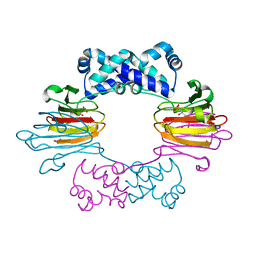 | | Fe(II)-HppE with R-HPP | | Descriptor: | Epoxidase, FE (II) ION, [(2R)-2-hydroxypropyl]phosphonic acid | | Authors: | Drennan, C.L. | | Deposit date: | 2011-06-07 | | Release date: | 2011-07-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis of regiospecificity of a mononuclear iron enzyme in antibiotic fosfomycin biosynthesis.
J.Am.Chem.Soc., 133, 2011
|
|
3SCF
 
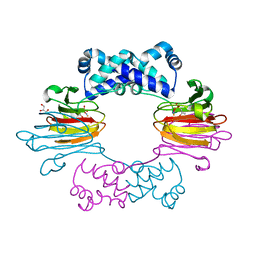 | | Fe(II)-HppE with S-HPP and NO | | Descriptor: | (S)-2-HYDROXYPROPYLPHOSPHONIC ACID, Epoxidase, FE (II) ION, ... | | Authors: | Drennan, C.L. | | Deposit date: | 2011-06-07 | | Release date: | 2011-07-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural basis of regiospecificity of a mononuclear iron enzyme in antibiotic fosfomycin biosynthesis.
J.Am.Chem.Soc., 133, 2011
|
|
4J1W
 
 | |
4J1X
 
 | |
4M7T
 
 | | Crystal structure of BtrN in complex with AdoMet and 2-DOIA | | Descriptor: | (1R,2S,3S,4R,5S)-5-aminocyclohexane-1,2,3,4-tetrol, BtrN, GLYCEROL, ... | | Authors: | Drennan, C.L, Goldman, P.J. | | Deposit date: | 2013-08-12 | | Release date: | 2013-10-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | X-ray analysis of butirosin biosynthetic enzyme BtrN redefines structural motifs for AdoMet radical chemistry.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4M7S
 
 | |
6OWR
 
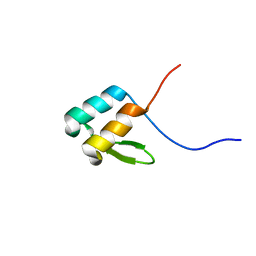 | | NMR solution structure of YfiD | | Descriptor: | Autonomous glycyl radical cofactor | | Authors: | Bowman, S.E.J, Drennan, C.L. | | Deposit date: | 2019-05-10 | | Release date: | 2019-07-10 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure and biochemical characterization of a spare part protein that restores activity to an oxygen-damaged glycyl radical enzyme.
J.Biol.Inorg.Chem., 24, 2019
|
|
4XC8
 
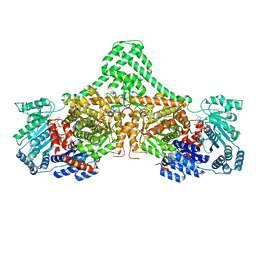 | | Isobutyryl-CoA mutase fused with bound butyryl-CoA, GDP, and Mg and without cobalamin (apo-IcmF/GDP) | | Descriptor: | Butyryl Coenzyme A, GUANOSINE-5'-DIPHOSPHATE, Isobutyryl-CoA mutase fused, ... | | Authors: | Jost, M, Drennan, C.L. | | Deposit date: | 2014-12-17 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Visualization of a radical B12 enzyme with its G-protein chaperone.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4XHM
 
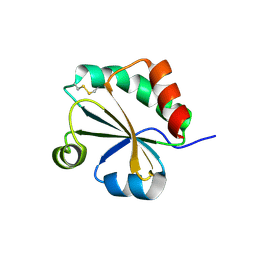 | |
5FAW
 
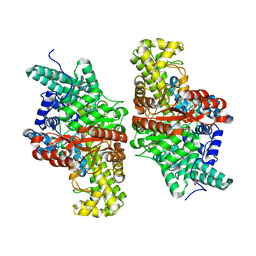 | | T502A mutant of choline TMA-lyase | | Descriptor: | CHOLINE ION, Choline trimethylamine-lyase, MALONATE ION, ... | | Authors: | Funk, M.A, Drennan, C.L. | | Deposit date: | 2015-12-12 | | Release date: | 2016-09-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.852 Å) | | Cite: | Molecular Basis of C-N Bond Cleavage by the Glycyl Radical Enzyme Choline Trimethylamine-Lyase.
Cell Chem Biol, 23, 2016
|
|
5FAU
 
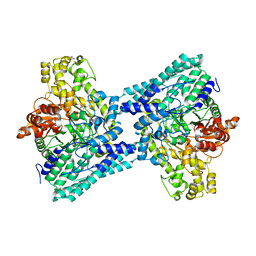 | | wild-type choline TMA lyase in complex with choline | | Descriptor: | CHOLINE ION, Choline trimethylamine-lyase, GLYCEROL | | Authors: | Funk, M.A, Drennan, C.L. | | Deposit date: | 2015-12-12 | | Release date: | 2016-09-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular Basis of C-N Bond Cleavage by the Glycyl Radical Enzyme Choline Trimethylamine-Lyase.
Cell Chem Biol, 23, 2016
|
|
8DPB
 
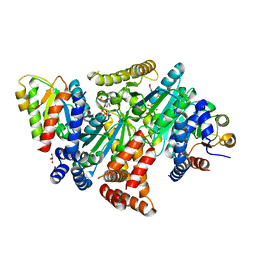 | | MeaB in complex with the cobalamin-binding domain of its target mutase with GMPPCP bound | | Descriptor: | GLYCEROL, MAGNESIUM ION, Methylmalonyl-CoA mutase accessory protein, ... | | Authors: | Vaccaro, F.A, Born, D.A, Drennan, C.L. | | Deposit date: | 2022-07-15 | | Release date: | 2023-03-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Structure of metallochaperone in complex with the cobalamin-binding domain of its target mutase provides insight into cofactor delivery.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
1ZZ6
 
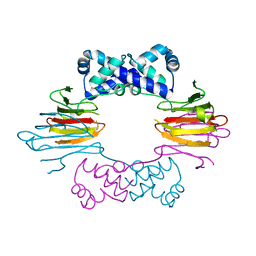 | | Crystal Structure of Apo-HppE | | Descriptor: | Hydroxypropylphosphonic Acid Epoxidase | | Authors: | Higgins, L.J, Yan, F, Liu, P, Liu, H.W, Drennan, C.L. | | Deposit date: | 2005-06-13 | | Release date: | 2005-07-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insight into antibiotic fosfomycin biosynthesis by a mononuclear iron enzyme
Nature, 437, 2005
|
|
1ZZ9
 
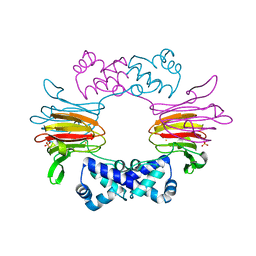 | | Crystal Structure of FeII HppE | | Descriptor: | FE (II) ION, Hydroxypropylphosphonic Acid Epoxidase, SULFATE ION | | Authors: | Higgins, L.J, Yan, F, Liu, P, Liu, H.W, Drennan, C.L. | | Deposit date: | 2005-06-13 | | Release date: | 2005-07-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insight into antibiotic fosfomycin biosynthesis by a mononuclear iron enzyme
Nature, 437, 2005
|
|
1ZZ7
 
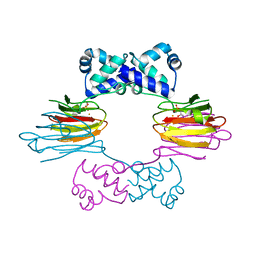 | | Crystal Structure of FeII HppE in Complex with Substrate form 1 | | Descriptor: | (S)-2-HYDROXYPROPYLPHOSPHONIC ACID, FE (II) ION, Hydroxyprophylphosphonic Acid Epoxidase | | Authors: | Higgins, L.J, Yan, F, Liu, P, Liu, H.W, Drennan, C.L. | | Deposit date: | 2005-06-13 | | Release date: | 2005-07-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insight into antibiotic fosfomycin biosynthesis by a mononuclear iron enzyme
Nature, 437, 2005
|
|
4XC7
 
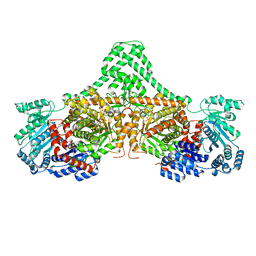 | |
4XC6
 
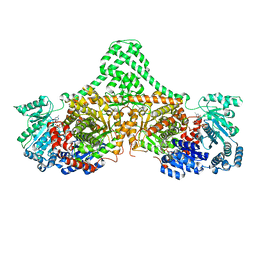 | | Isobutyryl-CoA mutase fused with bound adenosylcobalamin, GDP, and Mg (holo-IcmF/GDP) | | Descriptor: | 5'-DEOXYADENOSINE, ACETATE ION, COBALAMIN, ... | | Authors: | Jost, M, Drennan, C.L. | | Deposit date: | 2014-12-17 | | Release date: | 2015-02-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Visualization of a radical B12 enzyme with its G-protein chaperone.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
8V9I
 
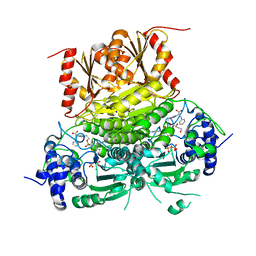 | | 1-deoxy-D-xylulose 5-phosphate synthase (DXPS) from Deinococcus radiodurans with D-phenylalanine-derived triazole acetylphosphonate (D-PheTrAP) bound | | Descriptor: | 1-deoxy-D-xylulose-5-phosphate synthase, 2-HYDROXY BUTANE-1,4-DIOL, 3-[(4-amino-2-methylpyrimidin-5-yl)methyl]-2-{(1S)-1-[(S)-(2-{1-[(1R)-1-carboxy-2-phenylethyl]-1H-1,2,3-triazol-4-yl}ethoxy)(hydroxy)phosphoryl]-1-hydroxyethyl}-5-(2-{[(S)-hydroxy(phosphonooxy)phosphoryl]oxy}ethyl)-4-methyl-1,3-thiazol-3-ium, ... | | Authors: | Chen, P.Y.-T, Drennan, C.L. | | Deposit date: | 2023-12-08 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.981 Å) | | Cite: | Potent Inhibition of E. coli DXP Synthase by a gem -Diaryl Bisubstrate Analog.
Acs Infect Dis., 10, 2024
|
|
5T6O
 
 | |
