8E4C
 
 | | IgM BCR fab truncated form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, B-cell antigen receptor complex-associated protein alpha chain,Yellow fluorescent protein, B-cell antigen receptor complex-associated protein beta chain, ... | | Authors: | Dong, Y, Pi, X, Wu, H, Reth, M. | | Deposit date: | 2022-08-18 | | Release date: | 2022-11-16 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural principles of B cell antigen receptor assembly.
Nature, 612, 2022
|
|
1P9E
 
 | | Crystal Structure Analysis of Methyl Parathion Hydrolase from Pseudomonas sp WBC-3 | | Descriptor: | CADMIUM ION, Methyl Parathion Hydrolase, POTASSIUM ION, ... | | Authors: | Dong, Y, Sun, L, Bartlam, M, Rao, Z, Zhang, X. | | Deposit date: | 2003-05-11 | | Release date: | 2004-05-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure Analysis of Methyl Parathion Hydrolase from Pseudomonas sp WBC-3
To be Published
|
|
8JP0
 
 | | structure of human sodium-calciumexchanger NCX1 | | Descriptor: | 2-{4-[(2,5-difluorophenyl)methoxy]phenoxy}-5-ethoxyaniline, Sodium/calcium exchanger 1 | | Authors: | Dong, Y, Zhao, Y. | | Deposit date: | 2023-06-09 | | Release date: | 2024-01-03 | | Last modified: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural insight into the allosteric inhibition of human sodium-calcium exchanger NCX1 by XIP and SEA0400.
Embo J., 43, 2024
|
|
6BY7
 
 | | Folding DNA into a lipid-conjugated nano-barrel for controlled reconstitution of membrane proteins | | Descriptor: | DNA (26-MER), DNA (27-MER), DNA (29-MER), ... | | Authors: | Dong, Y, Chen, S, Zhang, S, Sodroski, J, Yang, Z, Liu, D, Mao, Y. | | Deposit date: | 2017-12-20 | | Release date: | 2018-02-28 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (7.5 Å) | | Cite: | Folding DNA into a Lipid-Conjugated Nanobarrel for Controlled Reconstitution of Membrane Proteins.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
7DSV
 
 | | Structure of a human NHE1-CHP1 complex under pH 6.5 | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, Calcineurin B homologous protein 1, Sodium/hydrogen exchanger 1 | | Authors: | Dong, Y, Gao, Y, Li, B, Zhang, X.C, Zhao, Y. | | Deposit date: | 2021-01-03 | | Release date: | 2021-06-23 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure and mechanism of the human NHE1-CHP1 complex.
Nat Commun, 12, 2021
|
|
7DSW
 
 | | Structure of a human NHE1-CHP1 complex under pH 7.5 | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, Sodium/hydrogen exchanger 1 | | Authors: | Dong, Y, Gao, Y, Li, B, Zhang, X.C, Zhao, Y. | | Deposit date: | 2021-01-03 | | Release date: | 2021-06-23 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure and mechanism of the human NHE1-CHP1 complex.
Nat Commun, 12, 2021
|
|
7DSX
 
 | | Structure of a human NHE1-CHP1 complex under pH 7.5, bound by cariporide | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, Calcineurin B homologous protein 1, N-[bis(azanyl)methylidene]-3-methylsulfonyl-4-propan-2-yl-benzamide, ... | | Authors: | Dong, Y, Gao, Y, Li, B, Zhang, X.C, Zhao, Y. | | Deposit date: | 2021-01-03 | | Release date: | 2021-06-23 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure and mechanism of the human NHE1-CHP1 complex.
Nat Commun, 12, 2021
|
|
7VFS
 
 | | Human N-type voltage gated calcium channel CaV2.2-alpha2/delta1-beta1 complex, apo state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Dong, Y, Gao, Y, Wang, Y, Zhao, Y. | | Deposit date: | 2021-09-13 | | Release date: | 2021-11-03 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Closed-state inactivation and pore-blocker modulation mechanisms of human Ca V 2.2.
Cell Rep, 37, 2021
|
|
7VFV
 
 | | Human N-type voltage gated calcium channel CaV2.2-alpha2/delta1-beta1 complex, bound to PD173212 | | Descriptor: | (2~{S})-~{N}-[(2~{S})-1-(~{tert}-butylamino)-1-oxidanylidene-3-(4-phenylmethoxyphenyl)propan-2-yl]-2-[(4-~{tert}-butylphenyl)methyl-methyl-amino]-4-methyl-pentanamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Dong, Y, Gao, Y, Wang, Y, Zhao, Y. | | Deposit date: | 2021-09-13 | | Release date: | 2021-11-03 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Closed-state inactivation and pore-blocker modulation mechanisms of human Ca V 2.2.
Cell Rep, 37, 2021
|
|
7VFU
 
 | | Human N-type voltage gated calcium channel CaV2.2-alpha2/delta1-beta1 complex, bound to ziconotide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Dong, Y, Gao, Y, Wang, Y, Zhao, Y. | | Deposit date: | 2021-09-13 | | Release date: | 2021-11-03 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Closed-state inactivation and pore-blocker modulation mechanisms of human Ca V 2.2.
Cell Rep, 37, 2021
|
|
7VFW
 
 | | Human N-type voltage gated calcium channel CaV2.2-alpha2/delta1-beta1 complex, bound to CaV2.2-blocker1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-(4-chlorophenyl)-1-(2-methoxyphenyl)-3-(2,2,6,6-tetramethyloxan-4-yl)pyrazole, CALCIUM ION, ... | | Authors: | Dong, Y, Gao, Y, Wang, Y, Zhao, Y. | | Deposit date: | 2021-09-13 | | Release date: | 2021-11-03 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Closed-state inactivation and pore-blocker modulation mechanisms of human Ca V 2.2.
Cell Rep, 37, 2021
|
|
7X2U
 
 | | Structure of a human NHE3-CHP1 complex in the autoinhibited state | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, Calcineurin B homologous protein 1, Sodium/hydrogen exchanger 3, ... | | Authors: | Dong, Y, Li, H, Gao, Y, Zhang, X.C, Zhao, Y. | | Deposit date: | 2022-02-26 | | Release date: | 2022-04-20 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of a human NHE3-CHP1 complex in the autoinhibited state
Sci Adv, 2022
|
|
4RLW
 
 | | Crystal Structure of (3R)-hydroxyacyl-ACP dehydratase HadAB hetero-dimer from Mycobacterium tuberculosis complexed with Butein | | Descriptor: | (2E)-1-(2,4-dihydroxyphenyl)-3-(3,4-dihydroxyphenyl)prop-2-en-1-one, (3R)-hydroxyacyl-ACP dehydratase subunit HadA, (3R)-hydroxyacyl-ACP dehydratase subunit HadB, ... | | Authors: | Li, J, Dong, Y, Rao, Z.H. | | Deposit date: | 2014-10-18 | | Release date: | 2015-10-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.196 Å) | | Cite: | Molecular basis for the inhibition of beta-hydroxyacyl-ACP dehydratase HadAB complex from Mycobacterium tuberculosis by flavonoid inhibitors.
Protein Cell, 6, 2015
|
|
4RLU
 
 | | Crystal Structure of (3R)-hydroxyacyl-ACP dehydratase HadAB hetero-dimer from Mycobacterium tuberculosis complexed with 2',4,4'-trihydroxychalcone | | Descriptor: | (3R)-hydroxyacyl-ACP dehydratase subunit HadA, (3R)-hydroxyacyl-ACP dehydratase subunit HadB, 2',4,4'-TRIHYDROXYCHALCONE, ... | | Authors: | Li, J, Dong, Y, Rao, Z.H. | | Deposit date: | 2014-10-18 | | Release date: | 2015-10-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.198 Å) | | Cite: | Molecular basis for the inhibition of beta-hydroxyacyl-ACP dehydratase HadAB complex from Mycobacterium tuberculosis by flavonoid inhibitors.
Protein Cell, 6, 2015
|
|
4RLT
 
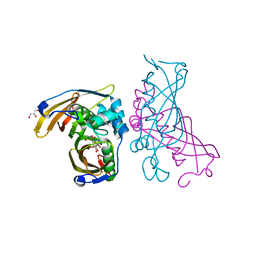 | | Crystal Structure of (3R)-hydroxyacyl-ACP dehydratase HadAB hetero-dimer from Mycobacterium tuberculosis complexed with Fisetin | | Descriptor: | (3R)-hydroxyacyl-ACP dehydratase subunit HadA, (3R)-hydroxyacyl-ACP dehydratase subunit HadB, 3,7,3',4'-TETRAHYDROXYFLAVONE, ... | | Authors: | Li, J, Dong, Y, Rao, Z.H. | | Deposit date: | 2014-10-18 | | Release date: | 2015-10-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.049 Å) | | Cite: | Molecular basis for the inhibition of beta-hydroxyacyl-ACP dehydratase HadAB complex from Mycobacterium tuberculosis by flavonoid inhibitors.
Protein Cell, 6, 2015
|
|
4RLJ
 
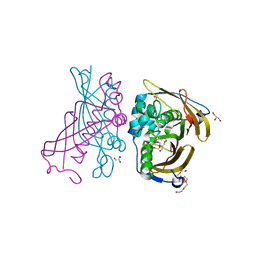 | | Crystal Structure of (3R)-hydroxyacyl-ACP dehydratase HadAB hetero-dimer from Mycobacterium tuberculosis | | Descriptor: | (3R)-hydroxyacyl-ACP dehydratase subunit HadA, (3R)-hydroxyacyl-ACP dehydratase subunit HadB, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, ... | | Authors: | Li, J, Dong, Y, Rao, Z.H. | | Deposit date: | 2014-10-17 | | Release date: | 2015-10-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Molecular basis for the inhibition of beta-hydroxyacyl-ACP dehydratase HadAB complex from Mycobacterium tuberculosis by flavonoid inhibitors.
Protein Cell, 6, 2015
|
|
8SV1
 
 | |
5W3M
 
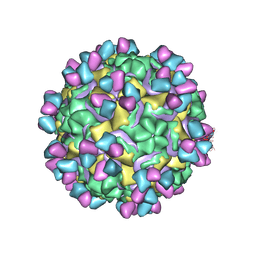 | | CryoEM structure of rhinovirus B14 in complex with C5 Fab (33 degrees Celsius, molar ratio 1:1, full particle) | | Descriptor: | C5 antibody variable heavy domain, C5 antibody variable light domain, viral protein 1, ... | | Authors: | Liu, Y, Dong, Y, Rossmann, M.G. | | Deposit date: | 2017-06-08 | | Release date: | 2017-07-12 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (2.26 Å) | | Cite: | Antibody-induced uncoating of human rhinovirus B14.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5W3O
 
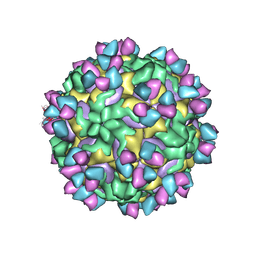 | | CryoEM structure of rhinovirus B14 in complex with C5 Fab (33 degrees Celsius, molar ratio 1:3, empty particle) | | Descriptor: | C5 antibody variable heavy domain, C5 antibody variable light domain, viral protein 1, ... | | Authors: | Liu, Y, Dong, Y, Rossmann, M.G. | | Deposit date: | 2017-06-08 | | Release date: | 2017-07-12 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Antibody-induced uncoating of human rhinovirus B14.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5W3L
 
 | | CryoEM structure of rhinovirus B14 in complex with C5 Fab (4 degrees Celsius, molar ratio 1:3, full particle) | | Descriptor: | C5 antibody variable heavy domain, C5 antibody variable light domain, viral protein 1, ... | | Authors: | Liu, Y, Dong, Y, Rossmann, M.G. | | Deposit date: | 2017-06-08 | | Release date: | 2017-07-12 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | Antibody-induced uncoating of human rhinovirus B14.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5W3E
 
 | | CryoEM structure of rhinovirus B14 in complex with C5 Fab (33 degrees Celsius, molar ratio 1:3, full particle) | | Descriptor: | C5 antibody variable heavy domain, C5 antibody variable light domain, viral protein 1, ... | | Authors: | Liu, Y, Dong, Y, Rossmann, M.G. | | Deposit date: | 2017-06-07 | | Release date: | 2017-07-12 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (2.53 Å) | | Cite: | Antibody-induced uncoating of human rhinovirus B14.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5YIM
 
 | | Structure of a Legionella effector | | Descriptor: | SdeA | | Authors: | Feng, Y, Dong, Y, Wang, W. | | Deposit date: | 2017-10-05 | | Release date: | 2018-05-30 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.394 Å) | | Cite: | Structural basis of ubiquitin modification by the Legionella effector SdeA.
Nature, 557, 2018
|
|
5YIK
 
 | |
5XX0
 
 | |
7SKC
 
 | |
