3J3X
 
 | |
5A2T
 
 | | The Molecular Basis for Flexibility in the Flexible Filamentous Plant Viruses | | Descriptor: | BAMBOO MOSAIC VIRUS, COAT PROTEIN | | Authors: | DiMaio, F, Chen, C.C, Yu, X, Frenz, B, Hsu, Y.H, Lin, N.S, Egelman, E.H. | | Deposit date: | 2015-05-23 | | Release date: | 2015-07-22 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | The Molecular Basis for Flexibility in the Flexible Filamentous Plant Viruses.
Nat.Struct.Mol.Biol., 22, 2015
|
|
3J9X
 
 | | A Virus that Infects a Hyperthermophile Encapsidates A-Form DNA | | Descriptor: | DNA, coat protein | | Authors: | DiMaio, F, Yu, X, Rensen, E, Krupovic, M, Prangishvili, D, Egelman, E. | | Deposit date: | 2015-03-21 | | Release date: | 2015-06-03 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | A virus that infects a hyperthermophile encapsidates A-form DNA.
Science, 348, 2015
|
|
5CW9
 
 | | Crystal structure of De novo designed ferredoxin-ferredoxin domain insertion protein | | Descriptor: | De novo designed ferredoxin-ferredoxin domain insertion protein | | Authors: | DiMaio, F, King, I.C, Gleixner, J, Doyle, L, Stoddard, B, Baker, D. | | Deposit date: | 2015-07-28 | | Release date: | 2015-09-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.108 Å) | | Cite: | Precise assembly of complex beta sheet topologies from de novo designed building blocks.
Elife, 4, 2015
|
|
6VPT
 
 | |
8UB3
 
 | | DpHF7 filament | | Descriptor: | DpHF7 filament | | Authors: | Lynch, E.M, Farrell, D, Shen, H, Kollman, J.M, DiMaio, F, Baker, D. | | Deposit date: | 2023-09-22 | | Release date: | 2024-04-10 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | De novo design of pH-responsive self-assembling helical protein filaments.
Nat Nanotechnol, 19, 2024
|
|
4RZP
 
 | | Crystal Structure of Engineered Protein. Northeast Structural Genomics Consortium (NESG) Target OR366. | | Descriptor: | Engineered Protein OR366 | | Authors: | Vorobiev, S, Parmeggiani, F, Dimaio, F, Seetharaman, J, Sahdev, S, Xiao, R, Kogan, S, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2014-12-23 | | Release date: | 2015-01-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.804 Å) | | Cite: | Crystal Structure of Engineered Protein OR366
To be Published
|
|
7RSL
 
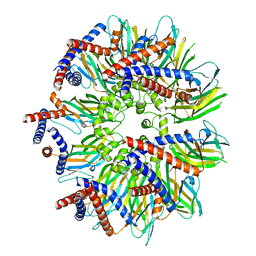 | | Seipin forms a flexible cage at lipid droplet formation sites | | Descriptor: | Seipin | | Authors: | Arlt, H, Sui, X, Folger, B, Adams, C, Chen, X, Remme, R, Hamprecht, F.A, DiMaio, F, Liao, M, Goodman, J.M, Farese Jr, R.V, Walther, T.C. | | Deposit date: | 2021-08-11 | | Release date: | 2022-02-09 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Seipin forms a flexible cage at lipid droplet formation sites.
Nat.Struct.Mol.Biol., 29, 2022
|
|
8DIT
 
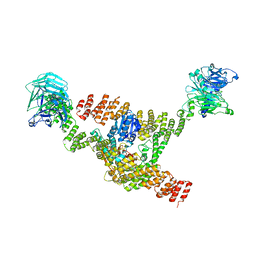 | | Cryo-EM structure of a HOPS core complex containing Vps33, Vps16, and Vps18 | | Descriptor: | Vacuolar protein sorting-associated protein 16, Vacuolar protein sorting-associated protein 18, Vacuolar protein sorting-associated protein 33 | | Authors: | Port, S.A, Farrell, P.D, Jeffrey, P.D, DiMaio, F, Hughson, F.M. | | Deposit date: | 2022-06-29 | | Release date: | 2022-08-31 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (5.1 Å) | | Cite: | Cryo-EM structure of the HOPS core complex and its implication for SNARE assembly
To Be Published
|
|
5SZS
 
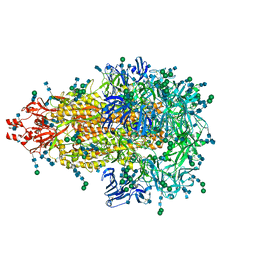 | | Glycan shield and epitope masking of a coronavirus spike protein observed by cryo-electron microscopy | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Walls, A.C, Tortorici, M.A, Frenz, B, Snijder, J, Li, W, Rey, F.A, DiMaio, F, Bosch, B.J, Veesler, D. | | Deposit date: | 2016-08-15 | | Release date: | 2016-09-14 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Glycan shield and epitope masking of a coronavirus spike protein observed by cryo-electron microscopy.
Nat.Struct.Mol.Biol., 23, 2016
|
|
4TQL
 
 | | Computationally designed three helix bundle | | Descriptor: | Three helix bundle | | Authors: | Nannenga, B.L, Oberdorfer, G, DiMaio, F, Baker, D, Gonen, T. | | Deposit date: | 2014-06-11 | | Release date: | 2014-10-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | High thermodynamic stability of parametrically designed helical bundles.
Science, 346, 2014
|
|
5EC0
 
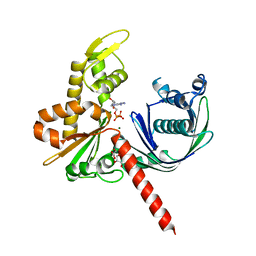 | | Crystal Structure of Actin-like protein Alp7A | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Alp7A, GLYCEROL, ... | | Authors: | Petek, N.A, Kraemer, J.A, Mullins, R.D, Agard, D.A, DiMaio, F, Baker, D. | | Deposit date: | 2015-10-20 | | Release date: | 2016-11-02 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and function of Alp7A reveal both conserved and unique features of plasmid segregation.
To Be Published
|
|
5OPY
 
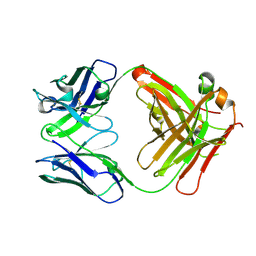 | | Crystal structure of anti-alphaVbeta3 integrin Fab LM609 | | Descriptor: | Heavy chain of LM609 Fab (antigen-binding fragment), Light chain of LM609 Fab (antigen-binding fragment) | | Authors: | Backovic, M, Veesler, D, Borst, A.J, James, Z.M, Zagotta, W, Ginsberg, M, Rey, F.A, DiMaio, F. | | Deposit date: | 2017-08-10 | | Release date: | 2017-10-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | The Therapeutic Antibody LM609 Selectively Inhibits Ligand Binding to Human alpha V beta 3 Integrin via Steric Hindrance.
Structure, 25, 2017
|
|
5FLU
 
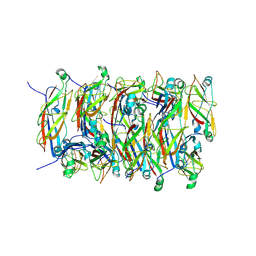 | | Structure of a Chaperone-Usher pilus reveals the molecular basis of rod uncoilin | | Descriptor: | PAP FIMBRIAL MAJOR PILIN PROTEIN | | Authors: | Hospenthal, M.K, Redzej, A, Dodson, K, Ukleja, M, Frenz, B, Hultgren, S.J, DiMaio, F, Egelman, E.H, Waksman, G. | | Deposit date: | 2015-10-28 | | Release date: | 2016-01-13 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of a Chaperone-Usher Pilus Reveals the Molecular Basis of Rod Uncoiling.
Cell(Cambridge,Mass.), 164, 2016
|
|
3SQF
 
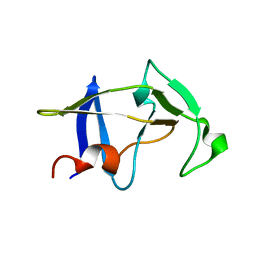 | | Crystal structure of monomeric M-PMV retroviral protease | | Descriptor: | Protease | | Authors: | Jaskolski, M, Kazmierczyk, M, Gilski, M, Krzywda, S, Pichova, I, Zabranska, H, Khatib, F, DiMaio, F, Cooper, S, Thompson, J, Popovic, Z, Baker, D, Group, Foldit Contenders | | Deposit date: | 2011-07-05 | | Release date: | 2011-09-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6324 Å) | | Cite: | Crystal structure of a monomeric retroviral protease solved by protein folding game players.
Nat.Struct.Mol.Biol., 18, 2011
|
|
6Y5V
 
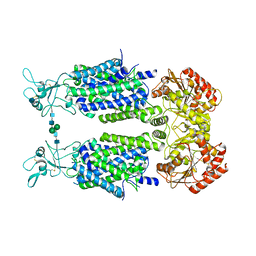 | | Structure of Human Potassium Chloride Transporter KCC3b (S45D/T940D/T997D) in KCl | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Solute carrier family 12 member 6, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Chi, G, Man, H, Ebenhoch, R, Reggiano, G, Pike, A.C.W, Wang, D, McKinley, G, Mukhopadhyay, S.M.M, MacLean, E.M, Chalk, R, Moreau, C, Snee, M, Bohstedt, T, Singh, N.K, Abrusci, P, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Marsden, B.D, Burgess-Brown, N.A, DiMaio, F, Duerr, K.L. | | Deposit date: | 2020-02-25 | | Release date: | 2020-07-15 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.08 Å) | | Cite: | Phospho-regulation, nucleotide binding and ion access control in potassium-chloride cotransporters.
Embo J., 40, 2021
|
|
6Y5R
 
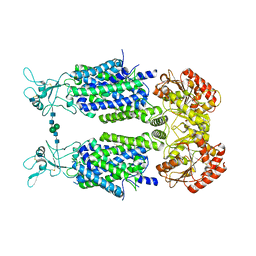 | | Structure of Human Potassium Chloride Transporter KCC3 S45D/T940D/T997D in NaCl | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Solute carrier family 12 member 6, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Chi, G, Man, H, Ebenhoch, R, Reggiano, G, Pike, A.C.W, Wang, D, McKinley, G, Mukhopadhyay, S.M.M, MacLean, B, Chalk, R, Moreau, C, Snee, M, Bohstedt, T, Singh, N.K, Abrusci, P, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Marsden, B.D, Burgess-Brown, N.A, DiMaio, F, Duerr, K.L, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-02-25 | | Release date: | 2020-03-11 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.76 Å) | | Cite: | Structure of Human Potassium Chloride Transporter KCC3 in NaCl
To be published
|
|
7RKH
 
 | | Yeast CTP Synthase (URA8) tetramer bound to ATP/UTP at neutral pH | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CTP synthase, MAGNESIUM ION, ... | | Authors: | Hansen, J.M, Lynch, E.M, Farrell, D.P, DiMaio, F, Quispe, J, Kollman, J.M. | | Deposit date: | 2021-07-22 | | Release date: | 2021-11-24 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM structures of CTP synthase filaments reveal mechanism of pH-sensitive assembly during budding yeast starvation.
Elife, 10, 2021
|
|
7RMF
 
 | | Substrate-bound Ura7 filament at low pH | | Descriptor: | CTP synthase | | Authors: | Hansen, J.M, Lynch, E.M, Farrell, D.P, DiMaio, F, Quispe, J, Kollman, J.M. | | Deposit date: | 2021-07-27 | | Release date: | 2021-11-24 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (7.3 Å) | | Cite: | Cryo-EM structures of CTP synthase filaments reveal mechanism of pH-sensitive assembly during budding yeast starvation.
Elife, 10, 2021
|
|
7RMV
 
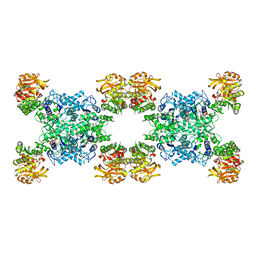 | | Yeast CTP Synthase (Ura7) H360R Filament bound to Substrates | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CTP synthase, URIDINE 5'-TRIPHOSPHATE | | Authors: | Hansen, J.M, Lynch, E.M, Farrell, D.P, DiMaio, F, Quispe, J, Kollman, J.M. | | Deposit date: | 2021-07-28 | | Release date: | 2021-11-24 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (6.7 Å) | | Cite: | Cryo-EM structures of CTP synthase filaments reveal mechanism of pH-sensitive assembly during budding yeast starvation.
Elife, 10, 2021
|
|
7RMK
 
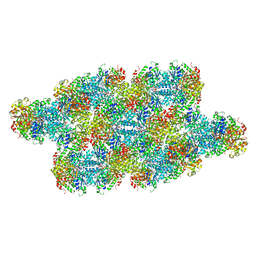 | | Yeast CTP Synthase (Ura7) Bundle bound to substrates at low pH | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CTP synthase, URIDINE 5'-TRIPHOSPHATE | | Authors: | Hansen, J.M, Lynch, E.M, Farrell, D.P, DiMaio, F, Quispe, J, Kollman, J.M. | | Deposit date: | 2021-07-27 | | Release date: | 2021-11-24 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Cryo-EM structures of CTP synthase filaments reveal mechanism of pH-sensitive assembly during budding yeast starvation.
Elife, 10, 2021
|
|
7RNR
 
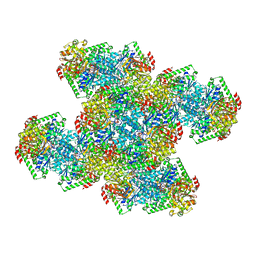 | | Yeast CTP Synthase (Ura8) Bundle Bound to Substrates at Low pH | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CTP synthase, MAGNESIUM ION, ... | | Authors: | Hansen, J.M, Lynch, E.M, Farrell, D.P, DiMaio, F, Quispe, J, Kollman, J.M. | | Deposit date: | 2021-07-29 | | Release date: | 2021-11-24 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures of CTP synthase filaments reveal mechanism of pH-sensitive assembly during budding yeast starvation.
Elife, 10, 2021
|
|
7RMC
 
 | | Yeast CTP Synthase (Ura7) filament bound to CTP at low pH | | Descriptor: | CTP synthase 1, CYTIDINE-5'-TRIPHOSPHATE | | Authors: | Hansen, J.M, Lynch, E.M, Farrell, D.P, DiMaio, F, Quispe, J, Kollman, J.M. | | Deposit date: | 2021-07-27 | | Release date: | 2021-11-24 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structures of CTP synthase filaments reveal mechanism of pH-sensitive assembly during budding yeast starvation.
Elife, 10, 2021
|
|
7RL5
 
 | | Yeast CTP Synthase (URA8) filament bound to CTP at low pH | | Descriptor: | CTP synthase, CYTIDINE-5'-TRIPHOSPHATE | | Authors: | Hansen, J.M, Lynch, E.M, Farrell, D.P, DiMaio, F, Quispe, J, Kollman, J.M. | | Deposit date: | 2021-07-23 | | Release date: | 2021-11-24 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structures of CTP synthase filaments reveal mechanism of pH-sensitive assembly during budding yeast starvation.
Elife, 10, 2021
|
|
7RNL
 
 | | Yeast CTP Synthase (Ura7) H360R Filament bound to Substrates | | Descriptor: | CTP synthase, CYTIDINE-5'-TRIPHOSPHATE, MAGNESIUM ION | | Authors: | Hansen, J.M, Lynch, E.M, Farrell, D.P, DiMaio, F, Quispe, J, Kollman, J.M. | | Deposit date: | 2021-07-29 | | Release date: | 2021-11-24 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structures of CTP synthase filaments reveal mechanism of pH-sensitive assembly during budding yeast starvation.
Elife, 10, 2021
|
|
