1QXZ
 
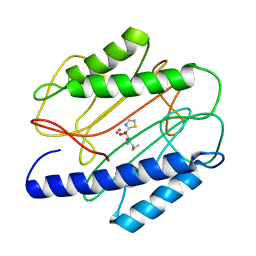 | | Crystal structure of S. aureus methionine aminopeptidase in complex with a ketoheterocycle inhibitor 119 | | Descriptor: | (2S)-2-AMINO-4-(METHYLSULFANYL)-1-(1,3-THIAZOL-2-YL)BUTANE-1,1-DIOL, COBALT (II) ION, methionyl aminopeptidase | | Authors: | Douangamath, A, Dale, G.E, D'Arcy, A, Oefner, C. | | Deposit date: | 2003-09-09 | | Release date: | 2004-03-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Crystal structures of staphylococcusaureus methionine aminopeptidase complexed with keto heterocycle and aminoketone inhibitors reveal the formation of a tetrahedral intermediate.
J.Med.Chem., 47, 2004
|
|
1QXW
 
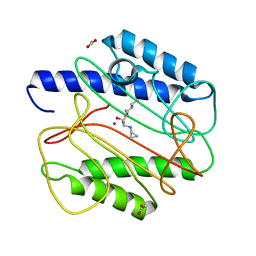 | | Crystal structure of Staphyloccocus aureus in complex with an aminoketone inhibitor 54135. | | Descriptor: | (3S)-3-AMINO-1-(CYCLOPROPYLAMINO)HEPTANE-2,2-DIOL, ACETATE ION, COBALT (II) ION, ... | | Authors: | Douangamath, A, Dale, G.E, D'Arcy, A, Oefner, C. | | Deposit date: | 2003-09-09 | | Release date: | 2004-03-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Crystal structures of staphylococcusaureus methionine aminopeptidase complexed with keto heterocycle and aminoketone inhibitors reveal the formation of a tetrahedral intermediate.
J.Med.Chem., 47, 2004
|
|
1QXY
 
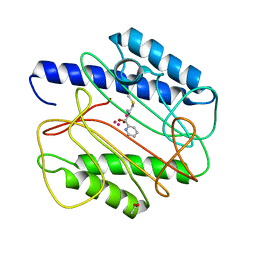 | | Crystal structure of S. aureus methionine aminopeptidase in complex with a ketoheterocycle 618 | | Descriptor: | (2S)-2-AMINO-4-(METHYLSULFANYL)-1-PYRIDIN-2-YLBUTANE-1,1-DIOL, ACETATE ION, COBALT (II) ION, ... | | Authors: | Douangamath, A, Dale, G.E, D'Arcy, A, Oefner, C. | | Deposit date: | 2003-09-09 | | Release date: | 2004-03-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Crystal structures of staphylococcusaureus methionine aminopeptidase complexed with keto heterocycle and aminoketone inhibitors reveal the formation of a tetrahedral intermediate.
J.Med.Chem., 47, 2004
|
|
1QFX
 
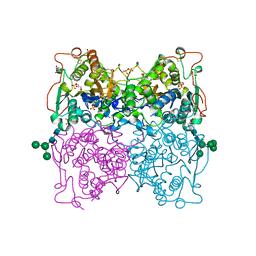 | | PH 2.5 ACID PHOSPHATASE FROM ASPERGILLUS NIGER | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, PROTEIN (PH 2.5 ACID PHOSPHATASE), ... | | Authors: | Kostrewa, D, Wyss, M, D'Arcy, A, Van Loon, A.P.G.M. | | Deposit date: | 1999-04-15 | | Release date: | 2000-04-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of Aspergillus niger pH 2.5 acid phosphatase at 2. 4 A resolution.
J.Mol.Biol., 288, 1999
|
|
2DHN
 
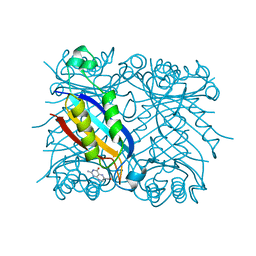 | | COMPLEX OF 7,8-DIHYDRONEOPTERIN ALDOLASE FROM STAPHYLOCOCCUS AUREUS WITH 6-HYDROXYMETHYL-7,8-DIHYDROPTERIN AT 2.2 A RESOLUTION | | Descriptor: | 2-AMINO-6-HYDROXYMETHYL-7,8-DIHYDRO-3H-PTERIDIN-4-ONE, 7,8-DIHYDRONEOPTERIN ALDOLASE | | Authors: | Hennig, M, D'Arcy, A, Hampele, I.C, Page, M.G.P, Oefner, C.H, Dale, G. | | Deposit date: | 1998-03-31 | | Release date: | 1999-04-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure and reaction mechanism of 7,8-dihydroneopterin aldolase from Staphylococcus aureus.
Nat.Struct.Biol., 5, 1998
|
|
1W0Y
 
 | | tf7a_3771 complex | | Descriptor: | 4-(4-BENZYLOXY-2-METHANESULFONYLAMINO-5-METHOXY-BENZYLAMINO)-BENZAMIDINE, BLOOD COAGULATION FACTOR VIIA, CACODYLATE ION, ... | | Authors: | Banner, D.W, D'Arcy, A, Groebke-Zbinden, K, Ackermann, J, Kirchhofer, D, Ji, Y.-H, Tschopp, T.B, Wallbaum, S, Weber, L. | | Deposit date: | 2004-05-27 | | Release date: | 2005-01-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Design of Selective Phenylglycine Amide Tissue Factor/Factor Viia Inhibitors
Bioorg.Med.Chem.Lett., 15, 2005
|
|
2TCL
 
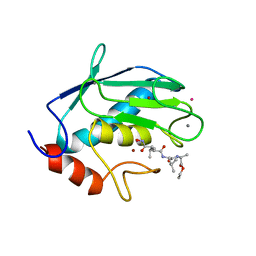 | | STRUCTURE OF THE CATALYTIC DOMAIN OF HUMAN FIBROBLAST COLLAGENASE COMPLEXED WITH AN INHIBITOR | | Descriptor: | CALCIUM ION, FIBROBLAST COLLAGENASE, SAMARIUM (III) ION, ... | | Authors: | Winkler, F.K, Borkakoti, N, D'Arcy, A. | | Deposit date: | 1994-09-07 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the catalytic domain of human fibroblast collagenase complexed with an inhibitor.
Nat.Struct.Biol., 1, 1994
|
|
1AD1
 
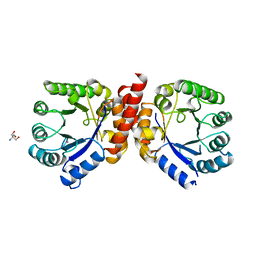 | | DIHYDROPTEROATE SYNTHETASE (APO FORM) FROM STAPHYLOCOCCUS AUREUS | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DIHYDROPTEROATE SYNTHETASE, POTASSIUM ION | | Authors: | Kostrewa, D, Oefner, C, D'Arcy, A. | | Deposit date: | 1997-02-19 | | Release date: | 1998-04-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and function of the dihydropteroate synthase from Staphylococcus aureus.
J.Mol.Biol., 268, 1997
|
|
1CBK
 
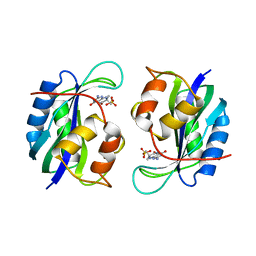 | | 7,8-DIHYDRO-6-HYDROXYMETHYLPTERIN-PYROPHOSPHOKINASE FROM HAEMOPHILUS INFLUENZAE | | Descriptor: | 7,8-DIHYDRO-7,7-DIMETHYL-6-HYDROXYPTERIN, PROTEIN (7,8-DIHYDRO-6-HYDROXYMETHYLPTERIN-PYROPHOSPHOKINASE), SULFATE ION | | Authors: | Hennig, M, D'Arcy, A, Dale, G, Oefner, C. | | Deposit date: | 1999-02-26 | | Release date: | 2000-03-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | The structure and function of the 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase from Haemophilus influenzae.
J.Mol.Biol., 287, 1999
|
|
1PFQ
 
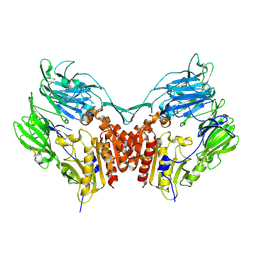 | | crystal structure of human apo dipeptidyl peptidase IV / CD26 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Dipeptidyl peptidase IV soluble form | | Authors: | Oefner, C, D'Arcy, A, Mac Sweeney, A, Pierau, S, Gardiner, R, Dale, G.E. | | Deposit date: | 2003-05-27 | | Release date: | 2003-07-01 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High-resolution structure of human apo dipeptidyl peptidase IV/CD26 and its complex with 1-[([2-[(5-iodopyridin-2-yl)amino]-ethyl]amino)-acetyl]-2-cyano-(S)-pyrrolidine.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1IYL
 
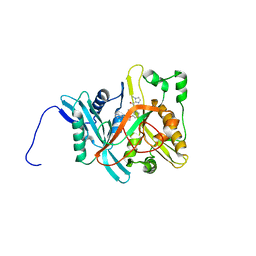 | | Crystal Structure of Candida albicans N-myristoyltransferase with Non-peptidic Inhibitor | | Descriptor: | (1-METHYL-1H-IMIDAZOL-2-YL)-(3-METHYL-4-{3-[(PYRIDIN-3-YLMETHYL)-AMINO]-PROPOXY}-BENZOFURAN-2-YL)-METHANONE, Myristoyl-CoA:Protein N-Myristoyltransferase | | Authors: | Sogabe, S, Fukami, T.A, Morikami, K, Shiratori, Y, Aoki, Y, D'Arcy, A, Winkler, F.K, Banner, D.W, Ohtsuka, T. | | Deposit date: | 2002-08-29 | | Release date: | 2002-12-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal Structures of Candida albicans N-Myristoyltransferase with Two Distinct Inhibitors
CHEM.BIOL., 9, 2002
|
|
1IYK
 
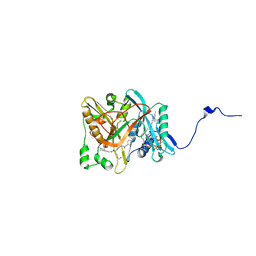 | | Crystal structure of candida albicans N-myristoyltransferase with myristoyl-COA and peptidic inhibitor | | Descriptor: | MYRISTOYL-COA:PROTEIN N-MYRISTOYLTRANSFERASE, TETRADECANOYL-COA, [CYCLOHEXYLETHYL]-[[[[4-[2-METHYL-1-IMIDAZOLYL-BUTYL]PHENYL]ACETYL]-SERYL]-LYSINYL]-AMINE | | Authors: | Sogabe, S, Fukami, T.A, Morikami, K, Shiratori, Y, Aoki, Y, D'Arcy, A, Winkler, F.K, Banner, D.W, Ohtsuka, T. | | Deposit date: | 2002-08-29 | | Release date: | 2002-12-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of Candida albicans N-Myristoyltransferase with Two Distinct Inhibitors
CHEM.BIOL., 9, 2002
|
|
1W2K
 
 | | tf7a_4380 complex | | Descriptor: | (2R)-2-({4-[AMINO(IMINO)METHYL]PHENYL}AMINO)-N-BENZYL-2-(3,4-DIMETHOXYPHENYL)ACETAMIDE, BLOOD COAGULATION FACTOR VIIA, CACODYLATE ION, ... | | Authors: | Banner, D.W, D'Arcy, A, Groebke-Zbinden, K, Ackermann, J, Kirchhofer, D, Ji, Y.-H, Tschopp, T.B, Wallbaum, S, Weber, L. | | Deposit date: | 2004-07-06 | | Release date: | 2005-06-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Design of Selective Phenylglycine Amide Tissue Factor/Factor Viia Inhibitors
Bioorg.Med.Chem.Lett., 15, 2005
|
|
4A92
 
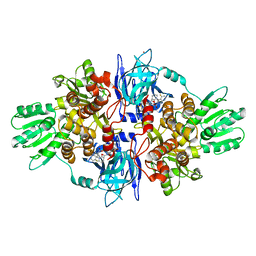 | | Full-length HCV NS3-4A protease-helicase in complex with a macrocyclic protease inhibitor. | | Descriptor: | (1'R,2R,2'S,6S,24AS)-17-FLUORO-6-(1-METHYL-2-OXOPIPERIDINE-3-CARBOXAMIDO)-19,19-DIOXIDO-5,21,24-TRIOXO-2'-VINYL-1,2,3,5,6,7,8,9,10,11,12,13,14,20,21,23,24,24A-OCTADECAHYDROSPIRO[BENZO[S]PYRROLO[2,1-G][1,2,5,8,18]THIATETRAAZACYCLOICOSINE-22,1'-CYCLOPRO-2-CARBOXYLATEPAN]-2-YL 4-FLUOROISOINDOLINE, SERINE PROTEASE NS3, ZINC ION | | Authors: | Schiering, N, D'Arcy, A, Simic, O, Eder, J, Raman, P, Svergun, D.I, Bodendorf, U. | | Deposit date: | 2011-11-23 | | Release date: | 2011-12-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | A Macrocyclic Hcv Ns3/4A Protease Inhibitor Interacts with Protease and Helicase Residues in the Complex with its Full- Length Target.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
1KLI
 
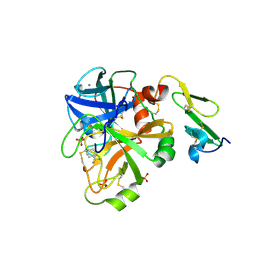 | | Cofactor-and substrate-assisted activation of factor VIIa | | Descriptor: | BENZAMIDINE, CALCIUM ION, GLYCEROL, ... | | Authors: | Sichler, K, Banner, D.W, D'Arcy, A, Hopfner, K.P, Huber, R, Bode, W, Kresse, G.B, Kopetzki, E, Brandstetter, H. | | Deposit date: | 2001-12-12 | | Release date: | 2002-09-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Crystal Structure of Uninhibited Factor VIIa Link its Cofactor and Substrate-assisted Activation to Specific Interactions
J.Mol.Biol., 322, 2002
|
|
1KLJ
 
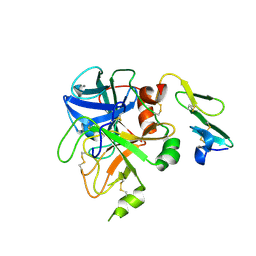 | | Crystal structure of uninhibited factor VIIa | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, factor VIIa | | Authors: | Sichler, K, Banner, D, D'Arcy, A, Hopfner, K.P, Huber, R, Bode, W, Kresse, G.B, Kopetzki, E, Brandstetter, H. | | Deposit date: | 2001-12-12 | | Release date: | 2002-10-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Crystal structures of uninhibited factor VIIa link its cofactor and substrate-assisted activation to specific interactions.
J.Mol.Biol., 322, 2002
|
|
2FP7
 
 | |
2FOM
 
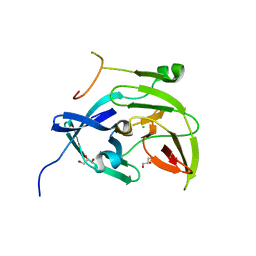 | | Dengue Virus NS2B/NS3 Protease | | Descriptor: | CHLORIDE ION, GLYCEROL, polyprotein | | Authors: | Schiering, N, Kroemer, M, Renatus, M, Erbel, P, D'Arcy, A. | | Deposit date: | 2006-01-13 | | Release date: | 2006-03-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis for the activation of flaviviral NS3 proteases from dengue and West Nile virus.
Nat.Struct.Mol.Biol., 13, 2006
|
|
1DMT
 
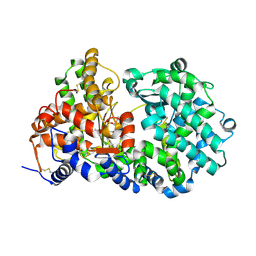 | | STRUCTURE OF HUMAN NEUTRAL ENDOPEPTIDASE COMPLEXED WITH PHOSPHORAMIDON | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, N-ALPHA-L-RHAMNOPYRANOSYLOXY(HYDROXYPHOSPHINYL)-L-LEUCYL-L-TRYPTOPHAN, ... | | Authors: | Oefner, C, D'Arcy, A, Hennig, M, Winkler, F.K, Dale, G.E. | | Deposit date: | 1999-12-15 | | Release date: | 2000-12-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of human neutral endopeptidase (Neprilysin) complexed with phosphoramidon.
J.Mol.Biol., 296, 2000
|
|
1DHN
 
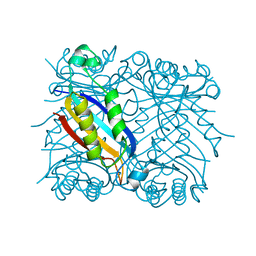 | | 1.65 ANGSTROM RESOLUTION STRUCTURE OF 7,8-DIHYDRONEOPTERIN ALDOLASE FROM STAPHYLOCOCCUS AUREUS | | Descriptor: | 7,8-DIHYDRONEOPTERIN ALDOLASE | | Authors: | Hennig, M, D'Arcy, A, Hampele, I.C, Page, M.G.P, Oefner, C.H, Dale, G. | | Deposit date: | 1998-03-31 | | Release date: | 1999-04-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure and reaction mechanism of 7,8-dihydroneopterin aldolase from Staphylococcus aureus.
Nat.Struct.Biol., 5, 1998
|
|
1FG9
 
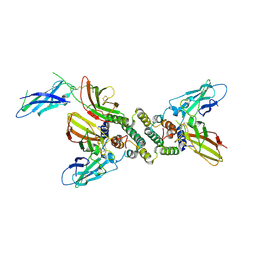 | | 3:1 COMPLEX OF INTERFERON-GAMMA RECEPTOR WITH INTERFERON-GAMMA DIMER | | Descriptor: | INTERFERON GAMMA, INTERFERON-GAMMA RECEPTOR ALPHA CHAIN | | Authors: | Thiel, D.J, le Du, M.-H, Walter, R.L, D'Arcy, A, Chene, C, Fountoulakis, M, Garotta, G, Winkler, F.K, Ealick, S.E. | | Deposit date: | 2000-07-28 | | Release date: | 2000-08-11 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Observation of an unexpected third receptor molecule in the crystal structure of human interferon-gamma receptor complex.
Structure Fold.Des., 8, 2000
|
|
1G97
 
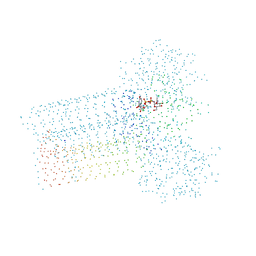 | | S.PNEUMONIAE GLMU COMPLEXED WITH UDP-N-ACETYLGLUCOSAMINE AND MG2+ | | Descriptor: | MAGNESIUM ION, N-ACETYLGLUCOSAMINE-1-PHOSPHATE URIDYLTRANSFERASE, SODIUM ION, ... | | Authors: | Kostrewa, D, D'Arcy, A, Kamber, M. | | Deposit date: | 2000-11-22 | | Release date: | 2001-05-22 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal structures of Streptococcus pneumoniae N-acetylglucosamine-1-phosphate uridyltransferase, GlmU, in apo form at 2.33 A resolution and in complex with UDP-N-acetylglucosamine and Mg(2+) at 1.96 A resolution.
J.Mol.Biol., 305, 2001
|
|
1FR6
 
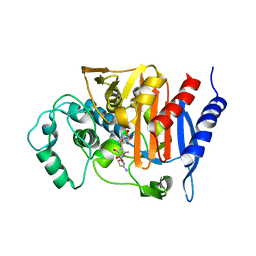 | | REFINED CRYSTAL STRUCTURE OF BETA-LACTAMASE FROM CITROBACTER FREUNDII INDICATES A MECHANISM FOR BETA-LACTAM HYDROLYSIS | | Descriptor: | 2-({[(1Z)-1-(2-amino-1,3-thiazol-4-yl)-2-oxo-2-{[(2S,3S)-1-oxo-3-(sulfoamino)butan-2-yl]amino}ethylidene]amino}oxy)-2-methylpropanoic acid, BETA-LACTAMASE | | Authors: | Oefner, C, D'Arcy, A, Daly, J.J, Winkler, F.K. | | Deposit date: | 2000-09-07 | | Release date: | 2001-01-17 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Refined crystal structure of beta-lactamase from Citrobacter freundii indicates a mechanism for beta-lactam hydrolysis.
Nature, 343, 1990
|
|
1G95
 
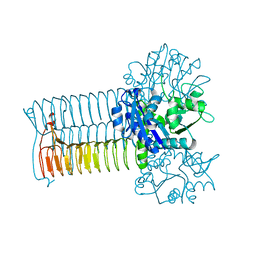 | | CRYSTAL STRUCTURE OF S.PNEUMONIAE GLMU, APO FORM | | Descriptor: | N-ACETYLGLUCOSAMINE-1-PHOSPHATE URIDYLTRANSFERASE | | Authors: | Kostrewa, D, D'Arcy, A, Kamber, M. | | Deposit date: | 2000-11-22 | | Release date: | 2001-05-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Crystal structures of Streptococcus pneumoniae N-acetylglucosamine-1-phosphate uridyltransferase, GlmU, in apo form at 2.33 A resolution and in complex with UDP-N-acetylglucosamine and Mg(2+) at 1.96 A resolution.
J.Mol.Biol., 305, 2001
|
|
1FR1
 
 | |
