5M5B
 
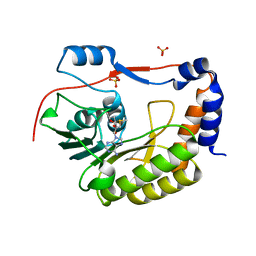 | | Crystal structure of Zika virus NS5 methyltransferase | | Descriptor: | CHLORIDE ION, GLYCEROL, NS5 methyltransferase, ... | | Authors: | Barral, K, Ortiz Lombardia, M, Coutard, B, Decroly, E, Lichiere, J. | | Deposit date: | 2016-10-21 | | Release date: | 2016-12-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Zika Virus Methyltransferase: Structure and Functions for Drug Design Perspectives.
J. Virol., 91, 2017
|
|
5IQ5
 
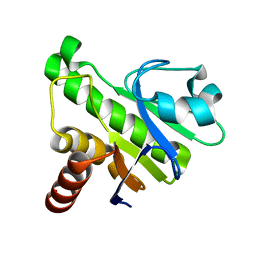 | | NMR solution structure of Mayaro virus macro domain | | Descriptor: | Macro domain | | Authors: | Melekis, E, Tsika, A.C, Bentrop, D, Papageorgiou, N, Coutard, B, Spyroulias, G.A. | | Deposit date: | 2016-03-10 | | Release date: | 2017-12-20 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Deciphering the Nucleotide and RNA Binding Selectivity of the Mayaro Virus Macro Domain.
J.Mol.Biol., 431, 2019
|
|
5ISN
 
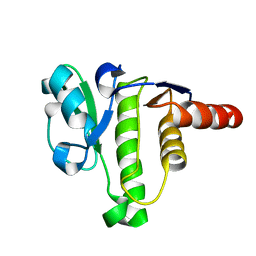 | | NMR solution structure of macro domain from Venezuelan equine encephalitis virus | | Descriptor: | Non-structural polyprotein | | Authors: | Makrynitsa, G.I, Ntonti, D, Marousis, K.D, Tsika, A.C, Papageorgiou, N, Coutard, B, Bentrop, D, Spyroulias, G.A. | | Deposit date: | 2016-03-15 | | Release date: | 2017-11-29 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Conformational plasticity of the VEEV macro domain is important for binding of ADP-ribose.
J.Struct.Biol., 206, 2019
|
|
3RUO
 
 | | Complex structure of HevB EV93 main protease 3C with Rupintrivir (AG7088) | | Descriptor: | 4-{2-(4-FLUORO-BENZYL)-6-METHYL-5-[(5-METHYL-ISOXAZOLE-3-CARBONYL)-AMINO]-4-OXO-HEPTANOYLAMINO}-5-(2-OXO-PYRROLIDIN-3-YL)-PENTANOIC ACID ETHYL ESTER, CHLORIDE ION, HEVB EV93 3C PROTEASE, ... | | Authors: | Kaczmarska, Z, Janowski, R, Costenaro, L, Coutard, B, Norder, H, Canard, B, Coll, M. | | Deposit date: | 2011-05-05 | | Release date: | 2011-09-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Basis for Antiviral Inhibition of the Main Protease, 3C, from Human Enterovirus 93.
J.Virol., 85, 2011
|
|
4UD1
 
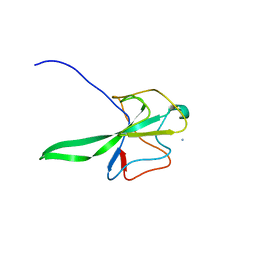 | | Structure of the N Terminal domain of the MERS CoV nucleocapsid | | Descriptor: | AMMONIUM ION, GLYCEROL, IMIDAZOLE, ... | | Authors: | Papageorgiou, N, Lichiere, J, Ferron, F, Canard, B, Coutard, B. | | Deposit date: | 2014-12-05 | | Release date: | 2015-12-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structural Characterization of the N-Terminal Part of the Mers-Cov Nucleocapsid by X-Ray Diffraction and Small-Angle X-Ray Scattering
Acta Crystallogr.,Sect.D, 72, 2016
|
|
5MQX
 
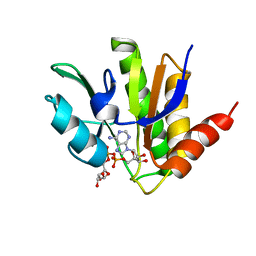 | | NMR solution structure of macro domain from Venezuelan equine encephalitis virus(VEEV) in complex with ADP-ribose | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Non-structural protein3 | | Authors: | Makrynitsa, G.I, Ntonti, D, Marousis, K.D, Matsoukas, M.T, Papageorgiou, N, Coutard, B, Bentrop, D, Spyroulias, G.A. | | Deposit date: | 2016-12-21 | | Release date: | 2018-07-04 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Conformational plasticity of the VEEV macro domain is important for binding of ADP-ribose.
J.Struct.Biol., 206, 2019
|
|
6G13
 
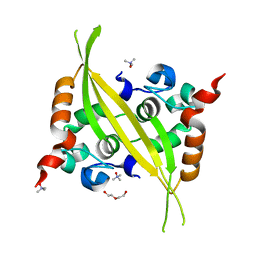 | | C-terminal domain of MERS-CoV nucleocapsid | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Nucleoprotein, ... | | Authors: | Nguyen, T.H.V, Ferron, F.P, Lichiere, J, Canard, B, Papageorgiou, N, Coutard, B. | | Deposit date: | 2018-03-20 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structure and oligomerization state of the C-terminal region of the Middle East respiratory syndrome coronavirus nucleoprotein.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
8RCQ
 
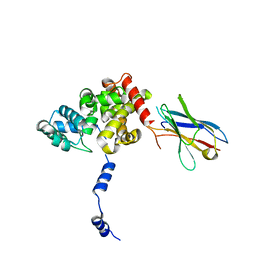 | | Structural flexibility of Nucleoprotein of the Toscana virus in the presence of a nanobody. | | Descriptor: | Nucleoprotein, VHH | | Authors: | Papageorgiou, N, Ferron, F, Coutard, B, Lichiere, J, Baklouti, A. | | Deposit date: | 2023-12-06 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structural flexibility of Toscana virus nucleoprotein in the presence of a single-chain camelid antibody.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
2CKW
 
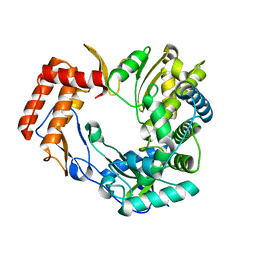 | | The 2.3 A resolution structure of the Sapporo virus RNA dependant RNA polymerase. | | Descriptor: | RNA-DIRECTED RNA POLYMERASE | | Authors: | Fullerton, S.W.B, Tucker, P.A, Rohayem, J, Coutard, B, Gebhardt, J, Gorbalenya, A, Canard, B. | | Deposit date: | 2006-04-24 | | Release date: | 2006-11-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Functional Characterization of Sapovirus RNA-Dependent RNA Polymerase.
J.Virol., 81, 2007
|
|
2H85
 
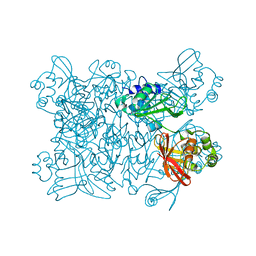 | | Crystal Structure of Nsp 15 from SARS | | Descriptor: | Putative orf1ab polyprotein | | Authors: | Ricagno, S, Egloff, M.P, Ulferts, R, Coutard, B, Nurizzo, D, Campanacci, V, Cambillau, C, Ziebuhr, J, Canard, B. | | Deposit date: | 2006-06-06 | | Release date: | 2006-08-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure and mechanistic determinants of SARS coronavirus nonstructural protein 15 define an endoribonuclease family.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
8CB3
 
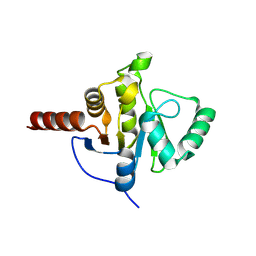 | | SARS-CoV Macro domain complexed with 3-(N-morpholino)propanesulfonic acid | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, GLYCEROL, Papain-like protease nsp3 | | Authors: | Morin, B, Ferron, F, Coutard, B, Canard, B, Marseilles Structural Genomics Program @ AFMB (MSGP) | | Deposit date: | 2023-01-25 | | Release date: | 2024-02-07 | | Last modified: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (1.572 Å) | | Cite: | Derivatives of MOPS: Promising scaffolds for SARS Coronaviruses macro domain-targeted inhibition
Febs J., 2025
|
|
4TU0
 
 | | CRYSTAL STRUCTURE OF CHIKUNGUNYA VIRUS NSP3 MACRO DOMAIN IN COMPLEX WITH A 2'-5' OLIGOADENYLATE TRIMER | | Descriptor: | 2'-5' OLIGOADENYLATE TRIMER, DI(HYDROXYETHYL)ETHER, Non-structural polyprotein 3 | | Authors: | Morin, B, Ferron, f.p, Malet, h, Coutard, b, Canard, b. | | Deposit date: | 2014-06-23 | | Release date: | 2014-07-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | CRYSTAL STRUCTURE OF CHIKUNGUNYA VIRUS NSP3 MACRO DOMAIN IN COMPLEX WITH A 2'-5' OLIGOADENYLATE TRIMER
TO BE PUBLISHED
|
|
8C0G
 
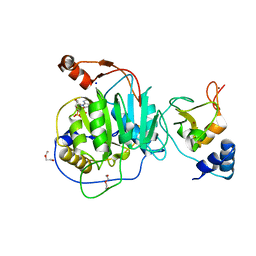 | | SARS-CoV nsp16-nsp10 complexed with N7-GTP | | Descriptor: | DI(HYDROXYETHYL)ETHER, Non-structural protein 7, SODIUM ION, ... | | Authors: | Ferron, F, Debarnot, C, Coutard, B, Canard, B, Marseilles Structural Genomics Program @ AFMB (MSGP) | | Deposit date: | 2022-12-16 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Observation of N7-GTP in nsp16-nsp10 Sars-CoV
To Be Published
|
|
6S3A
 
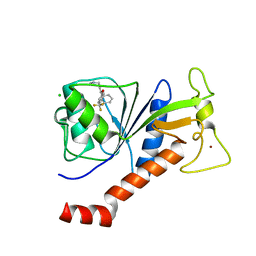 | | Coxsackie B3 2C protein in complex with S-Fluoxetine | | Descriptor: | (3S)-N-methyl-3-phenyl-3-[4-(trifluoromethyl)phenoxy]propan-1-amine, 2C protein, CHLORIDE ION, ... | | Authors: | El Kazzi, P, Papageorgiou, N, Ferron, F.P, Bauer, L, van Kuppeveld, F, Coutard, B. | | Deposit date: | 2019-06-24 | | Release date: | 2021-01-13 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Fluoxetine targets an allosteric site in the enterovirus 2C AAA+ ATPase and stabilizes a ring-shaped hexameric complex.
Sci Adv, 8, 2022
|
|
6T3W
 
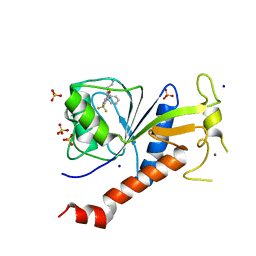 | | Coxsackie B3 2C protein in complex with S-Fluoxetine | | Descriptor: | (3S)-N-methyl-3-phenyl-3-[4-(trifluoromethyl)phenoxy]propan-1-amine, 2C protein, SODIUM ION, ... | | Authors: | El Kazzi, P, Papageorgiou, N, Ferron, F.P, Bauer, L, van Kuppeveld, F, Coutard, B. | | Deposit date: | 2019-10-11 | | Release date: | 2020-11-18 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Fluoxetine targets an allosteric site in the enterovirus 2C AAA+ ATPase and stabilizes a ring-shaped hexameric complex.
Sci Adv, 8, 2022
|
|
4U4I
 
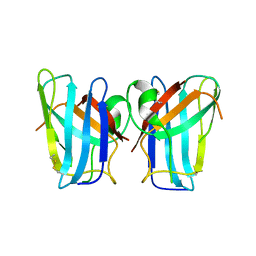 | | Megavirus chilensis superoxide dismutase | | Descriptor: | Cu/Zn superoxide dismutase | | Authors: | Lartigue, A, Claverie, J.-M, Burlat, B, Coutard, B, Abergel, C. | | Deposit date: | 2014-07-23 | | Release date: | 2014-11-05 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The megavirus chilensis cu,zn-superoxide dismutase: the first viral structure of a typical cellular copper chaperone-independent hyperstable dimeric enzyme.
J.Virol., 89, 2015
|
|
2XYQ
 
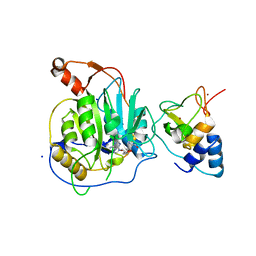 | | Crystal structure of the nsp16 nsp10 SARS coronavirus complex | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, NON-STRUCTURAL PROTEIN 10, ... | | Authors: | Decroly, E, Debarnot, C, Ferron, F, Bouvet, M, Coutard, B, Imbert, I, Gluais, L, Papageorgiou, N, Ortiz-Lombardia, M, Lescar, J, Canard, B. | | Deposit date: | 2010-11-18 | | Release date: | 2011-10-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure and Functional Analysis of the Sars-Coronavirus RNA CAP 2'-O-Methyltransferase Nsp10/Nsp16 Complex.
Plos Pathog., 7, 2011
|
|
2XYV
 
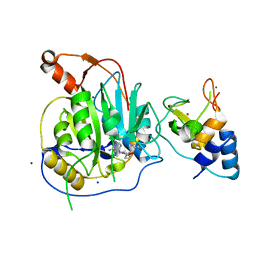 | | Crystal structure of the nsp16 nsp10 SARS coronavirus complex | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, NON-STRUCTURAL PROTEIN 10, ... | | Authors: | Decroly, E, Debarnot, C, Ferron, F, Bouvet, M, Coutard, B, Imbert, I, Gluais, L, Papageorgiou, N, Ortiz-Lombardia, M, Lescar, J, Canard, B. | | Deposit date: | 2010-11-19 | | Release date: | 2011-10-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Crystal Structure and Functional Analysis of the Sars-Coronavirus RNA CAP 2'-O-Methyltransferase Nsp10/Nsp16 Complex.
Plos Pathog., 7, 2011
|
|
2XYR
 
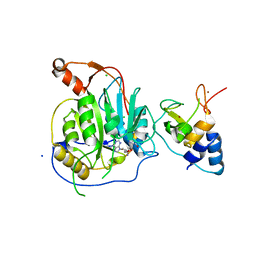 | | Crystal structure of the nsp16 nsp10 SARS coronavirus complex | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, NON-STRUCTURAL PROTEIN 10, ... | | Authors: | Decroly, E, Debarnot, C, Ferron, F, Bouvet, M, Coutard, B, Imbert, I, Gluais, L, Papageorgiou, N, Ortiz-Lombardia, M, Lescar, J, Canard, B. | | Deposit date: | 2010-11-18 | | Release date: | 2011-10-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure and Functional Analysis of the Sars-Coronavirus RNA CAP 2'-O-Methyltransferase Nsp10/Nsp16 Complex.
Plos Pathog., 7, 2011
|
|
3OV9
 
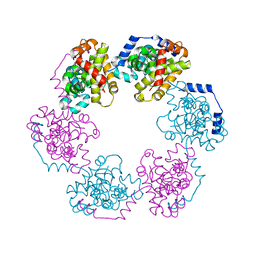 | | Structure of the Nucleoprotein from Rift Valley Fever Virus | | Descriptor: | NITRITE ION, Nucleoprotein, SODIUM ION | | Authors: | Ferron, F, Danek, E.I, Li, Z, Luo, D, Wong, Y.H, Coutard, B, Lantez, V, Charrel, R, Canard, B, Walz, T, Lescar, J. | | Deposit date: | 2010-09-16 | | Release date: | 2011-05-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The hexamer structure of Rift Valley fever virus nucleoprotein suggests a mechanism for its assembly into ribonucleoprotein complexes
Plos Pathog., 7, 2011
|
|
3OUO
 
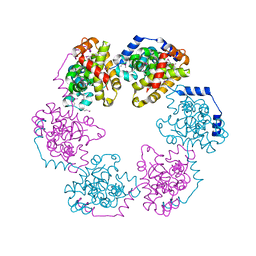 | | Structure of the Nucleoprotein from Rift Valley Fever Virus | | Descriptor: | NITRITE ION, Nucleoprotein | | Authors: | Ferron, F, Danek, E.I, Li, Z, Luo, D, Wong, Y.H, Coutard, B, Lantez, V, Charrel, R, Canard, B, Walz, T, Lescar, J. | | Deposit date: | 2010-09-15 | | Release date: | 2011-05-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The hexamer structure of Rift Valley fever virus nucleoprotein suggests a mechanism for its assembly into ribonucleoprotein complexes
Plos Pathog., 7, 2011
|
|
3Q3X
 
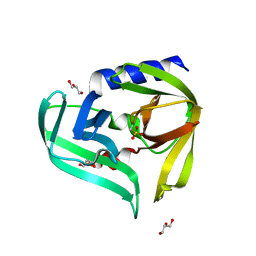 | | Crystal structure of the main protease (3C) from human enterovirus B EV93 | | Descriptor: | GLYCEROL, HEVB EV93 3C protease, MAGNESIUM ION | | Authors: | Costenaro, L, Sola, M, Coutard, B, Norder, H, Canard, B, Coll, M. | | Deposit date: | 2010-12-22 | | Release date: | 2011-09-07 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for Antiviral Inhibition of the Main Protease, 3C, from Human Enterovirus 93.
J.Virol., 85, 2011
|
|
3Q3Y
 
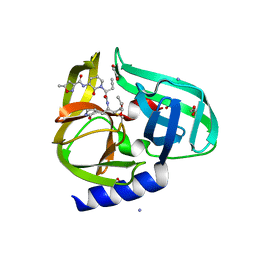 | | Complex structure of HEVB EV93 main protease 3C with Compound 1 (AG7404) | | Descriptor: | 1,2-ETHANEDIOL, AMMONIUM ION, HEVB EV93 3C protease, ... | | Authors: | Costenaro, L, Kaczmarska, Z, Arnan, C, Sola, M, Coutard, B, Norder, H, Canard, B, Coll, M. | | Deposit date: | 2010-12-22 | | Release date: | 2011-09-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Structural Basis for Antiviral Inhibition of the Main Protease, 3C, from Human Enterovirus 93.
J.Virol., 85, 2011
|
|
2W5E
 
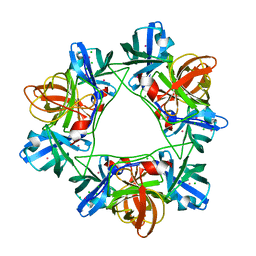 | | Structural and biochemical analysis of human pathogenic astrovirus serine protease at 2.0 Angstrom resolution | | Descriptor: | CADMIUM ION, CHLORIDE ION, PUTATIVE SERINE PROTEASE | | Authors: | Speroni, S, Rohayem, J, Nenci, S, Bonivento, D, Robel, I, Barthel, J, Coutard, B, Canard, B, Mattevi, A. | | Deposit date: | 2008-12-10 | | Release date: | 2009-03-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Biochemical Analysis of Human Pathogenic Astrovirus Serine Protease at 2.0 A Resolution.
J.Mol.Biol., 387, 2009
|
|
2V6J
 
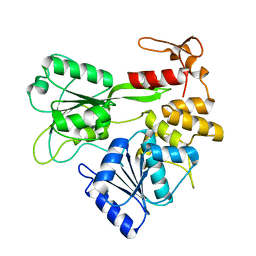 | | Kokobera Virus Helicase: Mutant Met47Thr | | Descriptor: | RNA HELICASE | | Authors: | Speroni, S, De Colibus, L, Coutard, B, Canard, B, Mattevi, A. | | Deposit date: | 2007-07-18 | | Release date: | 2008-03-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and Biochemical Analysis of Kokobera Virus Helicase
Proteins: Struct., Funct., Bioinf., 70, 2008
|
|
