6M32
 
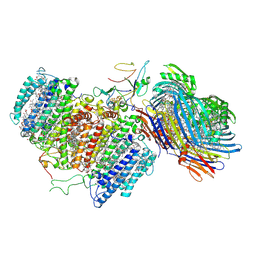 | | Cryo-EM structure of FMO-RC complex from green sulfur bacteria | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, 2-[(1E,3E,5E,7E,9E,11E,13E,15E,17E,19E)-3,7,12,16,20,24-hexamethylpentacosa-1,3,5,7,9,11,13,15,17,19,23-undecaenyl]-1,3,4-trimethyl-benzene, ... | | Authors: | Chen, J.H, Zhang, X. | | Deposit date: | 2020-03-02 | | Release date: | 2020-11-25 | | Last modified: | 2020-12-09 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Architecture of the photosynthetic complex from a green sulfur bacterium.
Science, 370, 2020
|
|
8GWA
 
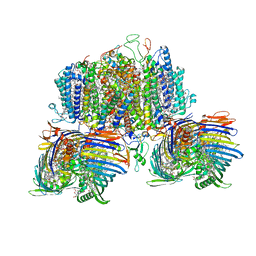 | | Structure of the intact photosynthetic light-harvesting antenna-reaction center complex from a green sulfur bacterium | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, 2-[(1E,3E,5E,7E,9E,11E,13E,15E,17E,19E)-3,7,12,16,20,24-hexamethylpentacosa-1,3,5,7,9,11,13,15,17,19,23-undecaenyl]-1,3,4-trimethyl-benzene, ... | | Authors: | Chen, J.H, Zhang, X. | | Deposit date: | 2022-09-16 | | Release date: | 2022-11-23 | | Last modified: | 2023-02-15 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-electron microscopy structure of the intact photosynthetic light-harvesting antenna-reaction center complex from a green sulfur bacterium.
J Integr Plant Biol, 65, 2023
|
|
7XR8
 
 | |
5YNZ
 
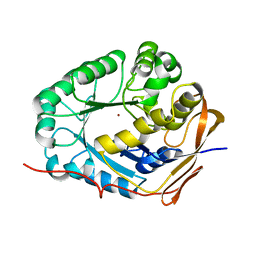 | | Crystal structure of the dihydroorotase domain (K1556A) of human CAD | | Descriptor: | CAD protein, ZINC ION | | Authors: | Huang, Y.H, Chen, K.L, Cheng, J.H, Huang, C.Y. | | Deposit date: | 2017-10-26 | | Release date: | 2018-10-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.774 Å) | | Cite: | Crystal structures of monometallic dihydropyrimidinase and the human dihydroorotase domain K1556A mutant reveal no lysine carbamylation within the active site
Biochem. Biophys. Res. Commun., 505, 2018
|
|
7DWQ
 
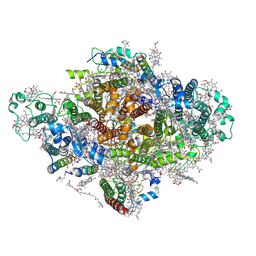 | | Photosystem I from a chlorophyll d-containing cyanobacterium Acaryochloris marina | | Descriptor: | (6'R,11cis,11'cis,13cis,15cis)-4',5'-didehydro-5',6'-dihydro-beta,beta-carotene, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Chen, J.H, Zhang, X, Shen, J.R. | | Deposit date: | 2021-01-17 | | Release date: | 2021-06-02 | | Last modified: | 2021-10-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | A unique photosystem I reaction center from a chlorophyll d-containing cyanobacterium Acaryochloris marina.
J Integr Plant Biol, 63, 2021
|
|
5V9U
 
 | | Crystal Structure of small molecule ARS-1620 covalently bound to K-Ras G12C | | Descriptor: | (S)-1-{4-[6-chloro-8-fluoro-7-(2-fluoro-6-hydroxyphenyl)quinazolin-4-yl] piperazin-1-yl}propan-1-one, CALCIUM ION, GLYCEROL, ... | | Authors: | Janes, M.R, Zhang, J, Li, L.-S, Hansen, R, Peters, U, Guo, X, Chen, Y, Babbar, A, Firdaus, S.J, Feng, J, Chen, J.H, Li, S, Brehmer, D, Darjania, L, Li, S, Long, Y.O, Thach, C, Liu, Y, Zarieh, A, Ely, T, Kucharski, J.M, Kessler, L.V, Wu, T, Wang, Y, Yao, Y, Deng, X, Zarrinkar, P, Dashyant, D, Lorenzi, M.V, Hu-Lowe, D, Patricelli, M.P, Ren, P, Liu, Y. | | Deposit date: | 2017-03-23 | | Release date: | 2018-02-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Targeting KRAS Mutant Cancers with a Covalent G12C-Specific Inhibitor.
Cell, 172, 2018
|
|
3SJM
 
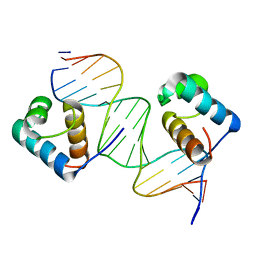 | | Crystal Structure Analysis of TRF2-Dbd-DNA complex | | Descriptor: | DNA (5'-D(*CP*TP*CP*TP*AP*GP*GP*GP*TP*TP*AP*GP*GP*GP*TP*TP*AP*G)-3'), DNA (5'-D(*TP*CP*TP*AP*AP*CP*CP*CP*TP*AP*AP*CP*CP*CP*TP*AP*GP*A)-3'), Telomeric repeat-binding factor 2 | | Authors: | Nair, S.K, Sliverman, S.K, Chen, J.H, Xiao, Y. | | Deposit date: | 2011-06-21 | | Release date: | 2012-10-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal Structure Analysis of TRF2-Dbd-DNA complex
To be Published
|
|
5F2E
 
 | | Crystal Structure of small molecule ARS-853 covalently bound to K-Ras G12C | | Descriptor: | 1-[3-[4-[2-[[4-chloranyl-5-(1-methylcyclopropyl)-2-oxidanyl-phenyl]amino]ethanoyl]piperazin-1-yl]azetidin-1-yl]prop-2-en-1-one, GLYCEROL, GLYCINE, ... | | Authors: | Patricelli, M.P, Janes, M.R, Li, L.-S, Hansen, R, Peters, U, Kessler, L.V, Chen, Y, Kucharski, J.M, Feng, J, Ely, T, Chen, J.H, Firdaus, S.J, Babbar, A, Ren, P, Liu, Y. | | Deposit date: | 2015-12-01 | | Release date: | 2016-01-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Selective Inhibition of Oncogenic KRAS Output with Small Molecules Targeting the Inactive State.
Cancer Discov, 6, 2016
|
|
6IIJ
 
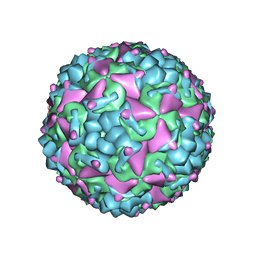 | | Cryo-EM structure of CV-A10 mature virion | | Descriptor: | SPHINGOSINE, VP1, VP2, ... | | Authors: | Chen, J.H, Ye, X.H, Cong, Y, Huang, Z. | | Deposit date: | 2018-10-06 | | Release date: | 2018-11-07 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Coxsackievirus A10 atomic structure facilitating the discovery of a broad-spectrum inhibitor against human enteroviruses.
Cell Discov, 5, 2019
|
|
5ZE8
 
 | | Crystal structure of a penta-heme cytochrome c552 from Thermochromatium tepidum | | Descriptor: | GLYCEROL, HEME C, SULFATE ION, ... | | Authors: | Yu, L.-J, Chen, J.-H, Shen, J.-R. | | Deposit date: | 2018-02-27 | | Release date: | 2018-04-25 | | Last modified: | 2019-10-02 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Properties and structure of a low-potential, penta-heme cytochrome c552from a thermophilic purple sulfur photosynthetic bacterium Thermochromatium tepidum.
Photosyn. Res., 139, 2019
|
|
3NKB
 
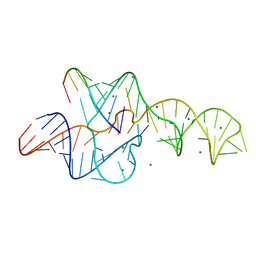 | | A 1.9A crystal structure of the HDV ribozyme precleavage suggests both Lewis acid and general acid mechanisms contribute to phosphodiester cleavage | | Descriptor: | DNA/RNA (5'-D(*(DUR))-D(*GP*G)-R(P*CP*UP*UP*GP*CP*A)-3'), MAGNESIUM ION, The hepatitis delta virus ribozyme | | Authors: | Chen, J.-H, Yajima, R, Chadalavada, D.M, Chase, E, Bevilacqua, P.C, Golden, B.L. | | Deposit date: | 2010-06-18 | | Release date: | 2010-09-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.916 Å) | | Cite: | A 1.9 A crystal structure of the HDV ribozyme precleavage suggests both Lewis acid and general acid mechanisms contribute to phosphodiester cleavage.
Biochemistry, 49, 2010
|
|
6IIO
 
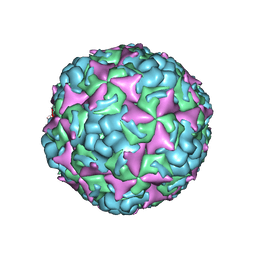 | | Cryo-EM structure of CV-A10 native empty particle | | Descriptor: | VP0, VP1, VP3 | | Authors: | Chen, J.H, Ye, X.H, Cong, Y, Huang, Z. | | Deposit date: | 2018-10-07 | | Release date: | 2018-11-07 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Coxsackievirus A10 atomic structure facilitating the discovery of a broad-spectrum inhibitor against human enteroviruses.
Cell Discov, 5, 2019
|
|
5YKD
 
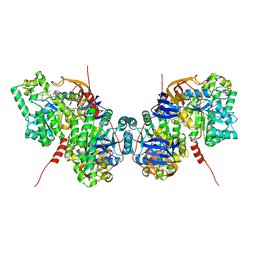 | |
6AJD
 
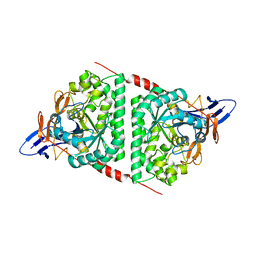 | |
7YMI
 
 | | PSII-Pcb Dimer of Acaryochloris Marina | | Descriptor: | (1R,2S)-4-{(1E,3E,5E,7E,9E,11E,13E,15E,17E)-18-[(4S)-4-hydroxy-2,6,6-trimethylcyclohex-1-en-1-yl]-3,7,12,16-tetramethyloctadeca-1,3,5,7,9,11,13,15,17-nonaen-1-yl}-2,5,5-trimethylcyclohex-3-en-1-ol, (6'R,11cis,11'cis,13cis,15cis)-4',5'-didehydro-5',6'-dihydro-beta,beta-carotene, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, ... | | Authors: | Shen, L.L, Gao, Y.Z, Wang, W.D, Zhang, X, Shen, J.R, Wang, P.Y, Han, G.Y. | | Deposit date: | 2022-07-28 | | Release date: | 2023-08-16 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of a unique PSII-Pcb tetrameric megacomplex in a chlorophyll d -containing cyanobacterium.
Sci Adv, 10, 2024
|
|
7YMM
 
 | | PSII-Pcb Tetramer of Acaryochloris Marina | | Descriptor: | (1R,2S)-4-{(1E,3E,5E,7E,9E,11E,13E,15E,17E)-18-[(4S)-4-hydroxy-2,6,6-trimethylcyclohex-1-en-1-yl]-3,7,12,16-tetramethyloctadeca-1,3,5,7,9,11,13,15,17-nonaen-1-yl}-2,5,5-trimethylcyclohex-3-en-1-ol, (6'R,11cis,11'cis,13cis,15cis)-4',5'-didehydro-5',6'-dihydro-beta,beta-carotene, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, ... | | Authors: | Shen, L.L, Gao, Y.Z, Wang, W.D, Zhang, X, Shen, J.R, Wang, P.Y, Han, G.Y. | | Deposit date: | 2022-07-28 | | Release date: | 2023-08-16 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure of a unique PSII-Pcb tetrameric megacomplex in a chlorophyll d -containing cyanobacterium.
Sci Adv, 10, 2024
|
|
2RKJ
 
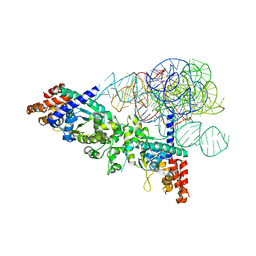 | | Cocrystal structure of a tyrosyl-tRNA synthetase splicing factor with a group I intron RNA | | Descriptor: | RNA (238-MER), RNA (5'-R(P*GP*CP*UP*U)-3'), Tyrosyl-tRNA synthetase | | Authors: | Paukstelis, P.J, Chen, J.-H, Chase, E, Lambowitz, A.M, Golden, B.L. | | Deposit date: | 2007-10-16 | | Release date: | 2008-01-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Structure of a tyrosyl-tRNA synthetase splicing factor bound to a group I intron RNA.
Nature, 451, 2008
|
|
4I3V
 
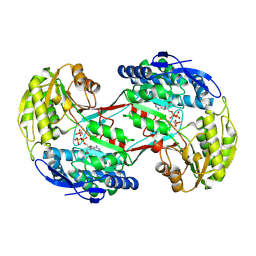 | |
4I3U
 
 | |
4I3W
 
 | |
4I3T
 
 | |
4I3X
 
 | |
4M6B
 
 | | Crystal structure of yeast Swr1-Z domain in complex with H2A.Z-H2B dimer | | Descriptor: | Chimera protein of Histone H2B.1 and Histone H2A.Z, Helicase SWR1 | | Authors: | Hong, J.J, Feng, H.Q, Wang, F, Ranjan, A, Chen, J.H, Jiang, J.S, Girlando, R, Xiao, T.S, Wu, C, Bai, Y.W. | | Deposit date: | 2013-08-09 | | Release date: | 2014-02-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | The Catalytic Subunit of the SWR1 Remodeler Is a Histone Chaperone for the H2A.Z-H2B Dimer.
Mol.Cell, 53, 2014
|
|
5YYU
 
 | |
4ETX
 
 | |
