5IQR
 
 | | Structure of RelA bound to the 70S ribosome | | Descriptor: | 30S ribosomal protein S10, 30S ribosomal protein S11, 30S ribosomal protein S12, ... | | Authors: | Brown, A, Fernandez, I.S, Gordiyenko, Y, Ramakrishnan, V. | | Deposit date: | 2016-03-11 | | Release date: | 2016-05-04 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Ribosome-dependent activation of stringent control.
Nature, 534, 2016
|
|
6MTB
 
 | | Rabbit 80S ribosome with P- and Z-site tRNAs (unrotated state) | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S10, ... | | Authors: | Brown, A, Baird, M.R, Yip, M.C.J, Murray, J, Shao, S. | | Deposit date: | 2018-10-19 | | Release date: | 2018-11-21 | | Last modified: | 2019-05-15 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structures of translationally inactive mammalian ribosomes.
Elife, 7, 2018
|
|
6MTC
 
 | | Rabbit 80S ribosome with Z-site tRNA and IFRD2 (unrotated state) | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S10, ... | | Authors: | Brown, A, Baird, M.R, Yip, M.C.J, Murray, J, Shao, S. | | Deposit date: | 2018-10-19 | | Release date: | 2018-11-21 | | Last modified: | 2019-05-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures of translationally inactive mammalian ribosomes.
Elife, 7, 2018
|
|
6MTD
 
 | | Rabbit 80S ribosome with eEF2 and SERBP1 (unrotated state with 40S head swivel) | | Descriptor: | 18S rRNA, 28S rRNA, 5.8S rRNA, ... | | Authors: | Brown, A, Baird, M.R, Yip, M.C.J, Murray, J, Shao, S. | | Deposit date: | 2018-10-19 | | Release date: | 2018-11-21 | | Last modified: | 2019-05-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structures of translationally inactive mammalian ribosomes.
Elife, 7, 2018
|
|
6MTE
 
 | | Rabbit 80S ribosome with eEF2 and SERBP1 (rotated state) | | Descriptor: | 18S rRNA, 28S rRNA, 5.8S rRNA, ... | | Authors: | Brown, A, Baird, M.R, Yip, M.C.J, Murray, J, Shao, S. | | Deposit date: | 2018-10-19 | | Release date: | 2018-11-21 | | Last modified: | 2019-05-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures of translationally inactive mammalian ribosomes.
Elife, 7, 2018
|
|
3J7Y
 
 | | Structure of the large ribosomal subunit from human mitochondria | | Descriptor: | 16S rRNA, ADENOSINE-5'-MONOPHOSPHATE, CRIF1, ... | | Authors: | Brown, A, Amunts, A, Bai, X.C, Sugimoto, Y, Edwards, P.C, Murshudov, G, Scheres, S.H.W, Ramakrishnan, V. | | Deposit date: | 2014-08-26 | | Release date: | 2014-10-15 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of the large ribosomal subunit from human mitochondria.
Science, 346, 2014
|
|
4CJM
 
 | | Crystal structure of human FGF18 | | Descriptor: | FIBROBLAST GROWTH FACTOR 18, SULFATE ION | | Authors: | Brown, A, Adam, L.E, Blundell, T.L. | | Deposit date: | 2013-12-21 | | Release date: | 2014-04-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Crystal Structure of Fibroblast Growth Factor 18 (Fgf18)
Protein Cell, 5, 2014
|
|
3DAR
 
 | |
3JAH
 
 | | Structure of a mammalian ribosomal termination complex with ABCE1, eRF1(AAQ), and the UAG stop codon | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 5.8S ribosomal RNA, ... | | Authors: | Brown, A, Shao, S, Murray, J, Hegde, R.S, Ramakrishnan, V. | | Deposit date: | 2015-06-10 | | Release date: | 2015-08-12 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Structural basis for stop codon recognition in eukaryotes.
Nature, 524, 2015
|
|
3JAG
 
 | | Structure of a mammalian ribosomal termination complex with ABCE1, eRF1(AAQ), and the UAA stop codon | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 5.8S ribosomal RNA, ... | | Authors: | Brown, A, Shao, S, Murray, J, Hegde, R.S, Ramakrishnan, V. | | Deposit date: | 2015-06-10 | | Release date: | 2015-08-12 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | Structural basis for stop codon recognition in eukaryotes.
Nature, 524, 2015
|
|
3JAI
 
 | | Structure of a mammalian ribosomal termination complex with ABCE1, eRF1(AAQ), and the UGA stop codon | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 5.8S ribosomal RNA, ... | | Authors: | Brown, A, Shao, S, Murray, J, Hegde, R.S, Ramakrishnan, V. | | Deposit date: | 2015-06-10 | | Release date: | 2015-08-12 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | Structural basis for stop codon recognition in eukaryotes.
Nature, 524, 2015
|
|
5OOM
 
 | | Structure of a native assembly intermediate of the human mitochondrial ribosome with unfolded interfacial rRNA | | Descriptor: | 16S ribosomal RNA, 39S ribosomal protein L10, mitochondrial, ... | | Authors: | Brown, A, Rathore, S, Kimanius, D, Aibara, S, Bai, X.C, Rorbach, J, Amunts, A, Ramakrishnan, V. | | Deposit date: | 2017-08-08 | | Release date: | 2017-09-13 | | Last modified: | 2023-03-15 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Structures of the human mitochondrial ribosome in native states of assembly.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5OOL
 
 | | Structure of a native assembly intermediate of the human mitochondrial ribosome with unfolded interfacial rRNA | | Descriptor: | 16S ribosomal RNA, 39S ribosomal protein L10, mitochondrial, ... | | Authors: | Brown, A, Rathore, S, Kimanius, D, Aibara, S, Bai, X.C, Rorbach, J, Amunts, A, Ramakrishnan, V. | | Deposit date: | 2017-08-08 | | Release date: | 2017-09-13 | | Last modified: | 2023-03-15 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Structures of the human mitochondrial ribosome in native states of assembly.
Nat. Struct. Mol. Biol., 24, 2017
|
|
9B4H
 
 | | Chlamydomonas reinhardtii mastigoneme filament | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, C-type lectin domain-containing protein, Tyrosine-protein kinase ephrin type A/B receptor-like domain-containing protein, ... | | Authors: | Dai, J, Ma, M, Zhang, R, Brown, A. | | Deposit date: | 2024-03-20 | | Release date: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Mastigoneme structure reveals insights into the O-linked glycosylation code of native hydroxyproline-rich helices.
Cell, 2024
|
|
7UNG
 
 | | 48-nm repeat of the human respiratory doublet microtubule | | Descriptor: | Cilia- and flagella-associated protein 161, Cilia- and flagella-associated protein 20, Cilia- and flagella-associated protein 45, ... | | Authors: | Gui, M, Croft, J.T, Zabeo, D, Acharya, V, Kollman, J.M, Burgoyne, T, Hoog, J.L, Brown, A. | | Deposit date: | 2022-04-11 | | Release date: | 2022-10-05 | | Last modified: | 2022-10-19 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | SPACA9 is a lumenal protein of human ciliary singlet and doublet microtubules.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7UN1
 
 | | 8-nm repeat of the human sperm tip singlet microtubule | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Gui, M, Croft, J.T, Zabeo, D, Acharya, V, Kollman, J.M, Burgoyne, T, Hoog, J.L, Brown, A. | | Deposit date: | 2022-04-08 | | Release date: | 2022-10-05 | | Last modified: | 2022-10-19 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | SPACA9 is a lumenal protein of human ciliary singlet and doublet microtubules.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8GLV
 
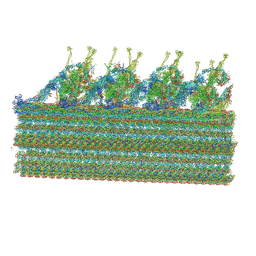 | |
7SQC
 
 | | Ciliary C1 central pair apparatus isolated from Chlamydomonas reinhardtii | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CPC1, Calmodulin, ... | | Authors: | Gui, M, Wang, X, Dutcher, S.K, Brown, A, Zhang, R. | | Deposit date: | 2021-11-05 | | Release date: | 2022-04-13 | | Last modified: | 2022-06-08 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Ciliary central apparatus structure reveals mechanisms of microtubule patterning.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7SOM
 
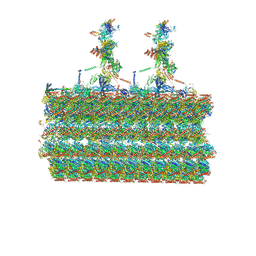 | | Ciliary C2 central pair apparatus isolated from Chlamydomonas reinhardtii | | Descriptor: | Cilia- and flagella-associated protein 20, FAP147, FAP178, ... | | Authors: | Gui, M, Wang, X, Dutcher, S.K, Brown, A, Zhang, R. | | Deposit date: | 2021-11-01 | | Release date: | 2022-04-13 | | Last modified: | 2022-06-08 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Ciliary central apparatus structure reveals mechanisms of microtubule patterning.
Nat.Struct.Mol.Biol., 29, 2022
|
|
8J07
 
 | |
7SHK
 
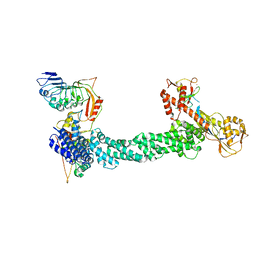 | | Structure of Xenopus laevis CRL2Lrr1 (State 1) | | Descriptor: | CULLIN_2 domain-containing protein, Elongin-C, Lrr1, ... | | Authors: | Zhou, H, Brown, A. | | Deposit date: | 2021-10-09 | | Release date: | 2021-12-08 | | Last modified: | 2021-12-29 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of CRL2Lrr1, the E3 ubiquitin ligase that promotes DNA replication termination in vertebrates.
Nucleic Acids Res., 49, 2021
|
|
7SHL
 
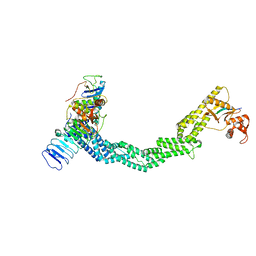 | | Structure of Xenopus laevis CRL2Lrr1 (State 2) | | Descriptor: | CULLIN_2 domain-containing protein, Elongin-C, Lrr1, ... | | Authors: | Zhou, H, Brown, A. | | Deposit date: | 2021-10-09 | | Release date: | 2021-12-08 | | Last modified: | 2021-12-29 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of CRL2Lrr1, the E3 ubiquitin ligase that promotes DNA replication termination in vertebrates.
Nucleic Acids Res., 49, 2021
|
|
6U3Q
 
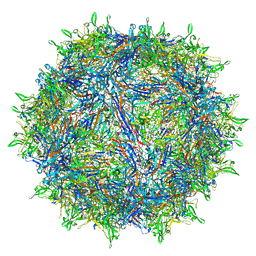 | | The atomic structure of a human adeno-associated virus capsid isolate (AAVhu69/AAVv66) | | Descriptor: | Capsid protein VP1 | | Authors: | Hsu, H.-L, Brown, A, Loveland, A, Tai, P, Korostelev, A, Gao, G. | | Deposit date: | 2019-08-22 | | Release date: | 2020-05-27 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.46 Å) | | Cite: | Structural characterization of a novel human adeno-associated virus capsid with neurotropic properties.
Nat Commun, 11, 2020
|
|
8F5P
 
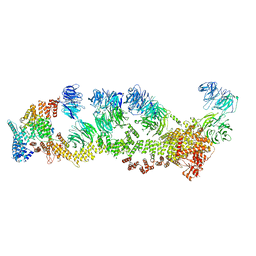 | | Structure of Leishmania tarentolae IFT-A (state 2) | | Descriptor: | Intraflagellar transport protein 122 homolog, Intraflagellar transport protein 122B, putative, ... | | Authors: | Zhou, H, Brown, A. | | Deposit date: | 2022-11-14 | | Release date: | 2022-12-21 | | Last modified: | 2023-01-11 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Mechanism of IFT-A polymerization into trains for ciliary transport.
Cell, 185, 2022
|
|
8F5O
 
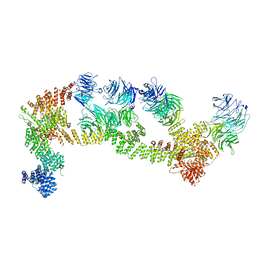 | | Structure of Leishmania tarentolae IFT-A (state 1) | | Descriptor: | Intraflagellar transport protein 122 homolog, Intraflagellar transport protein 122B, putative, ... | | Authors: | Zhou, H, Brown, A. | | Deposit date: | 2022-11-14 | | Release date: | 2022-12-21 | | Last modified: | 2023-01-11 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Mechanism of IFT-A polymerization into trains for ciliary transport.
Cell, 185, 2022
|
|
